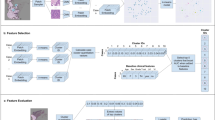
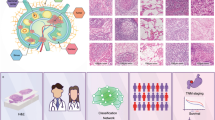
Abstract
Background
Identifying lymph node metastasis (LNM) relies mainly on indirect radiology. Current studies omitted the quantified associations with traits beyond cancer types, failing to provide generalisation performance across various tumour types.
Methods
4400 whole slide images across 11 cancer types were collected for training, cross-verification, and external validation of the pan-cancer lymph node metastasis (PC-LNM) model. We proposed an attention-based weakly supervised neural network based on self-supervised cancer-invariant features for the prediction task.
Results
PC-LNM achieved a test area under the curve (AUC) of 0.732 (95% confidence interval: 0.717–0.746, P < 0.0001) in fivefold cross-validation of multiple cancer types, which also demonstrated good generalisation in the external validation cohort with AUC of 0.699 (95% confidence interval: 0.658–0.737, P < 0.0001). The interpretability results derived from PC-LNM revealed that the regions with the highest attention scores identified by the model generally correspond to tumours with poorly differentiated morphologies. PC-LNM achieved superior performance over previously reported methods and could also act as an independent prognostic factor for patients across multiple tumour types.
Discussion
We presented an automated pan-cancer model for predicting the LNM status from primary tumour histology, which could act as a novel prognostic marker across multiple cancer types.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data supporting the findings of this study are available within the supplementary information and are also available from the authors upon reasonable request.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33.
Kawada K, Taketo MM. Significance and mechanism of lymph node metastasis in cancer progression. Cancer Res. 2011;71:1214–8.
Giuliano AE, Hunt KK, Ballman KV, Beitsch PD, Whitworth PW, et al. Axillary dissection vs no axillary dissection in women with invasive breast cancer and sentinel node metastasis: a randomized clinical trial. J Am Med Assoc. 2011;305:569–75.
Holten-Rossing H, Talman MM, Jylling AMB, Laenkholm AV, Kristensson M, Vainer B. Application of automated image analysis reduces the workload of manual screening of sentinel lymph node biopsies in breast cancer. Histopathology. 2017;71:866–73.
Fujimoto N, He Y, D’Addio M, Tacconi C, Detmar M, Dieterich LC. Single-cell mapping reveals new markers and functions of lymphatic endothelial cells in lymph nodes. PLoS Biol. 2020;18:e3000704.
Núñez NG, Tosello Boari J, Ramos RN, Richer W, Cagnard N, Anderfuhren CD, et al. Tumor invasion in draining lymph nodes is associated with Treg accumulation in breast cancer patients. Nat Commun. 2020;11:3272.
Nishiwada S, Sho M, Banwait JK, Yamamura K, Akahori T, Nakamura K, et al. A microRNA signature identifies pancreatic ductal adenocarcinoma patients at risk for lymph node metastases. Gastroenterology. 2020;159:562–74.
Barisoni L, Lafata KJ, Hewitt SM, Madabhushi A, Balis UGJ. Digital pathology and computational image analysis in nephropathology. Nat Rev Nephrol. 2020;16:669–85.
Yu F, Jung AW, Torne RV, Gonzalez S, Vhringer H, Shmatko A, et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat Cancer. 2020;1:800–10.
Wang X, Fang Y, Yang S, Zhu D, Wang M, Zhang J, et al. A hybrid network for automatic hepatocellular carcinoma segmentation in H&E-stained whole slide images. Med Image Anal. 2021;68:101914.
Lu MY, Williamson DFK, Chen TY, Chen RJ, Barbieri M, Mahmood F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Eng. 2021;5:555–70.
Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559–67.
Yamashita R, Long J, Longacre T, Peng L, Berry G, Martin B, et al. Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol. 2021;22:132–41.
Lu MY, Chen TY, Williamson DFK, Zhao M, Shady M, Lipkova J, et al. AI-based pathology predicts origins for cancers of unknown primary. Nature. 2021;594:106–10.
Hu Y, Su F, Dong K, Wang X, Zhao X, Jiang Y, et al. Deep learning system for lymph node quantification and metastatic cancer identification from whole-slide pathology images. Gastric Cancer. 2021;24:868–77.
Pham HHN, Futakuchi M, Bychkov A, Furukawa T, Kuroda K, Fukuoka J. Detection of lung cancer lymph node metastases from whole-slide histopathologic images using a two-step deep learning approach. Am J Pathol. 2019;189:2428–39.
Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. J Am Med Assoc. 2017;318:2199–210.
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–57.
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20.
Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, et al. Pathogenic germline variants in 10,389 adult cancers. Cell. 2018;173:355–70.
Ciga O, Xu T, Martel AL. Self supervised contrastive learning for digital histopathology. Mach Learn Appl. 2022;100198:1–14.
Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, et al. Swin transformer: hierarchical vision transformer using shifted windows. arXiv [Preprint] 2021. Available from: https://doi.org/10.48550/arXiv.2103.14030.
Wang X, Yang S, Zhang J, Wang M, Zhang J, Yang W, et al. Transformer-based unsupervised contrastive learning for histopathological image classification. Med Image Anal. 2022;81:102559.
DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45.
Shao Z, Bian H, Chen Y, Wang Y, Zhang J, Ji X, et al. TransMIL: transformer based correlated multiple instance learning for whole slide image classification. arXiv [Preprint] 2021. Available from: https://doi.org/10.48550/arXiv.2106.00908.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Lykke J, Roikjaer O, Jess P, Rosenberg J. Identification of risk factors associated with stage III disease in nonmetastatic colon cancer: results from a Prospective National Cohort Study. Ann Coloproctol. 2020;36:316–22.
Hanna AN, Sinnamon AJ, Roses RE, Kelz RR, Elder DE, Xu X, et al. Relationship between age and likelihood of lymph node metastases in patients with intermediate thickness melanoma (1.01-4.00 mm): a National Cancer Database study. J Am Acad Dermatol. 2019;80:433–40.
Glimelius B, Tiret E, Cervantes A, Arnold D. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24:vi81–8.
Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E, et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26:v8–30.
Brinker TJ, Kiehl L, Schmitt M, Jutzi TB, Krieghoff-Henning EI, Krahl D, et al. Deep learning approach to predict sentinel lymph node status directly from routine histology of primary melanoma tumours. Eur J Cancer. 2021;154:227–34.
Brockmoeller S, Echle A, Ghaffari Laleh N, Eiholm S, Malmstrøm ML, Plato Kuhlmann T, et al. Deep learning identifies inflamed fat as a risk factor for lymph node metastasis in early colorectal cancer. J Pathol. 2022;256:269–81.
Kwak MS, Lee HH, Yang JM, Cha JM, Jeon JW, Yoon JY, et al. Deep convolutional neural network-based lymph node metastasis prediction for colon cancer using histopathological images. Front Oncol. 2020;10:619803.
Zhao Y, Yang F, Fang Y, Liu H, Zhou N, Zhang J, et al. Predicting lymph node metastasis using histopathological images based on multiple instance learning with deep graph convolution. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2020. p. 4837–46.
Wang L, Jiao Y, Qiao Y, Zeng N, Yu R. A novel approach combined transfer learning and deep learning to predict TMB from histology image. Pattern Recognit Lett. 2020;135:244–8.
Acknowledgements
We appreciate the technical support for the professional pathology assessments in the comparison analysis from the department of Pathology, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine. Partial data used in this publication were retrieved from the National Cancer Institute Clinical Proteomic Tumour Analysis Consortium, the Cancer Genome Atlas, and the Cancer Imaging Archive used in this study.
Funding
This work was supported by the National Natural Science Foundation of China (81972393). The funding sources had no role in the design of the study; collection, analysis or interpretation of the data; writing of the report; the decision to submit for publication.
Author information
Authors and Affiliations
Contributions
JHZ, XH and JZ participated in the study conception and design, reviewed the paper and approved the final draft for submission. SC, JX and XW participated in the data collection, data analysis, wrote and approved the final draft for submission. SY and WY participated in data collection, reviewed the paper and approved the final draft for submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
No further ethical approval was required since all the slice images from the CPTAC cohort and the TCGA cohort were publicly available for research purposes.
Consent for publication
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, S., Xiang, J., Wang, X. et al. Deep learning-based pathology signature could reveal lymph node status and act as a novel prognostic marker across multiple cancer types. Br J Cancer 129, 46–53 (2023). https://doi.org/10.1038/s41416-023-02262-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02262-6
This article is cited by
-
Ein Blick in die Nachbardisziplin: eHealth in der Onkologie
Die Chirurgie (2024)
-
Novel research and future prospects of artificial intelligence in cancer diagnosis and treatment
Journal of Hematology & Oncology (2023)