Abstract
Background
Liquid biopsy is an alternative to tissue specimens for tumour genotyping. However, the frequency of genomic alterations with low circulating-tumour DNA (ctDNA) shedding is shown in pancreatic ductal adenocarcinoma (PDAC). We, therefore, investigated the prevalence of KRAS mutations and ctDNA fraction by the metastatic site in patients with PDAC.
Methods
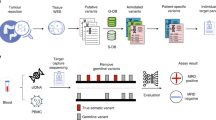
This study enrolled previously treated PDAC patients from a plasma genomic profiling study; ctDNA analysis was performed using Guardant360 at disease progression before initiating subsequent treatment.
Results
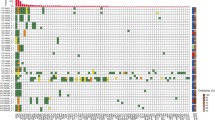
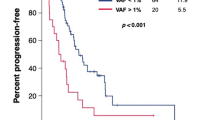
In 512 patients with PDAC, KRAS mutations were detected in 57%. The frequency of KRAS mutation in ctDNA differed depending on the metastatic organ; among patients with single-organ metastasis (n = 296), KRAS mutation detection rate was significantly higher in patients with metastasis to the liver (78%). In addition, the median maximum variant allele frequency (VAF) was higher with metastasis to the liver (1.9%) than with metastasis to the lungs, lymph nodes, peritoneum or with locally advanced disease (0.2%, 0.4%, 0.2% and 0.3%, respectively).
Conclusion
The prevalence of KRAS mutations and maximum VAF were higher in patients with metastasis to the liver than in those with metastasis to other sites. This study indicated the clinical utility of ctDNA analysis, especially in PDAC with liver metastases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The authors declare that all variant data used in the conduct of the analyses are available within the article and its Supplementary information. To protect the privacy and confidentiality of patients in this study, clinical data are not made publicly available in a repository or the supplementary material of the article but will be made available following reasonable request to the corresponding author.
References
Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Investig. 2012;122:639–53.
Singhi AD, George B, Greenbowe JR, Chung J, Suh J, Maitra A, et al. Real-time targeted genome profile analysis of pancreatic ductal adenocarcinomas identifies genetic alterations that might be targeted with existing drugs or used as biomarkers. Gastroenterology. 2019;156:2242–53.e4
Tempero MA. NCCN guidelines updates: pancreatic cancer. J Natl Compr Cancer Netw: JNCCN. 2019;17:603–5.
Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34.
Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13.
Sunami K, Ichikawa H, Kubo T, Kato M, Fujiwara Y, Shimomura A, et al. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: a hospital-based study. Cancer Sci. 2019;110:1480–90.
Zill OA, Greene C, Sebisanovic D, Siew LM, Leng J, Vu M, et al. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov. 2015;5:1040–8.
Nakamura Y, Taniguchi H, Ikeda M, Bando H, Kato K, Morizane C, et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med. 2020;26:1859–64.
Huerta M, Rosello S, Sabater L, Ferrer A, Tarazona N, Roda D, et al. Circulating tumor DNA detection by digital-droplet PCR in pancreatic ductal adenocarcinoma: a systematic review. Cancers. 2021;13:994.
Odegaard JI, Vincent JJ, Mortimer S, Vowles JV, Ulrich BC, Banks KC, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24:3539–49.
Bando H, Nakamura Y, Taniguchi H, Shiozawa M, Yasui H, Esaki T, et al. Impact of a metastatic site on circulating tumor DNA (ctDNA) analysis in patients (pts) with metastatic colorectal cancer (mCRC). J Clin Oncol. 2021;39:3554.
Ako S, Nouso K, Kinugasa H, Dohi C, Matushita H, Mizukawa S, et al. Utility of serum DNA as a marker for KRAS mutations in pancreatic cancer tissue. Pancreatology. 2017;17:285–90.
Watanabe F, Suzuki K, Tamaki S, Abe I, Endo Y, Takayama Y, et al. Longitudinal monitoring of KRAS-mutated circulating tumor DNA enables the prediction of prognosis and therapeutic responses in patients with pancreatic cancer. PLoS ONE. 2019;14:e0227366.
Patel H, Okamura R, Fanta P, Patel C, Lanman RB, Raymond VM, et al. Clinical correlates of blood-derived circulating tumor DNA in pancreatic cancer. J Hematol Oncol. 2019;12:130.
Zhang Q, Luo J, Wu S, Si H, Gao C, Xu W, et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov. 2020;10:1842–53.
Acknowledgements
The authors thank all of the patients and their families who participated in this study; all investigators and site personnel; Translational Research Support Section; Y Sakamoto and M Hata (National Cancer Center Hospital East) for data management; and all the National Cancer Center Hospital East Translational Research Support Section members.
Funding
This work was supported by SCRUM-Japan Funds (http://www.scrum-japan.ncc.go.jp/index.html).
Author information
Authors and Affiliations
Contributions
KU and YS contributed to the planning and conducting of studies, recruiting patients, acquisition of data, analysis and interpretation of the data and writing of the manuscript. MF, N Mizuno, KS, YK, TK, KO, NO, N Matsuhashi, SI, T Matsumoto, SS, T Otsuru, HH, H Okuyama, H Ohama, T Moriwaki and T Ohta contributed to the recruitment of patients and acquisition of data. JIO, YN, HB and TY contributed to the planning, conducting of studies and interpretation of data. MU, MI and CM contributed to the recruitment of patients, acquisition of data, planning and interpretation of data. All authors agree to be accountable for all aspects of the work and will ensure that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Competing interests
MF, KO, N Matsuhashi, SI, T Matsumoto, T Otsuru, HH, H Okuyama and H Ohama have nothing to disclose. KU reports honoraria from Chugai Pharmaceutical, Taiho Pharmaceutical and Yakult Honsha. YS reports honoraria from Takeda Pharmaceutical, Eli Lilly Japan, Chugai Pharmaceutical, Taiho Pharmaceutical, Bristol-Myers Squibb, Ono Pharmaceutical, Bayer, Daiichi Sankyo and Merck Biopharma; and grants from IQVIA and Parexel; and contributions or endowed chair from Takeda Pharmaceutical, Taiho Pharmaceutical, Chugai Pharmaceutical, Eli Lilly Japan, Sanofi and EN Otsuka Pharma. MU reports grants and personal fees from Taiho Pharmaceutical, AstraZeneca, Merck Biopharma, MSD, Ono Pharmaceutical, Incyte Corporation and Chugai Pharmaceutical; and personal fees from Nihon Servier; and grants from Astellas Pharma, Eisai and DFP. N Mizuno reports grants and personal fees from AstraZeneca, Novartis, Yakult Honsha, Ono Pharmaceutical, Taiho Pharmaceutical; and grants from MSD, Dainippon Sumitomo Pharma, ASLAN Pharmaceuticals, Incyte Corporation. and Seagen; and personal fees from Teijin Pharma, FUJIFILM Toyama Chemical, outside the submitted work. KS reports honoraria from Ono Pharmaceutical and Yakult Honsha; and grants (for the institution) from Bristol-Myers Squibb/Ono Pharmaceutical, Eisai and Incyte corporation. YK reports honoraria from Taiho Pharmaceutical, Incyte, Merck Biopharma, Yakult Honsha, and Eli Lilly; and grants from Takeda Pharmaceutical. TK reports honoraria from Taiho Pharmaceutical, Chugai Pharmaceutical, Ono Pharmaceutical and Eli Lilly. NO reports honoraria from Taiho Pharmaceutical, Eli Lilly, Eisai, Bayer Yakuhin, Chugai Pharmaceutical, Ono Pharmaceutical and Takeda Pharmaceutical; and advisory board from GlaxoSmithKline. SS reports grants from AstraZeneca, Incyte Corporation and Delta-Fly Pharma. T Moriwaki reports honoraria from Taiho Pharmaceutical, Eli Lilly Japan, Takeda Pharmaceutical, Chugai Pharmaceutical, Sanofi, Bayer Yakuhin, Merck Biopharma, Ono Pharmaceutical and Yakult Honsha; and grants from Taiho Pharmaceutical, MSD, Takeda Pharmaceutical, and Yakult Honsha. T Ohta reports honoraria from Chugai Pharmaceutical, Teijin Pharma, Takeda Pharmaceutical, Eisai, Yakult-Honsha, Daiichi Sankyo, Merck Biopharma, Ono Pharmaceutical, Taiho Pharmaceutical and Bristol-Myers Squibb; and grants from Takeda Pharmaceutical. JIO reports from, and stock interests in, Guardant Health. YN reports grants from Taiho Pharmaceutical, Chugai Pharmaceutical, Guardant Health, Genomedia, Daiichi Sankyo, Seagen, Roche Diagnostics. HB reports honoraria from Taiho Pharmaceutical, Eli Lilly Japan, Ono Pharmaceutical; and grants from Ono Pharmaceutical outside the submitted work. TY reports honoraria from Taiho Pharmaceutical, Chugai Pharmaceutical, Eli Lilly, Merck Biopharma, Bayer Yakuhin, Ono Pharmaceutical and MSD; and grants from Ono Pharmaceutical, Sanofi, Daiichi Sankyo, Parexel International, Pfizer Japan, Taiho Pharmaceutical, MSD, Amgen, Genomedia, Sysmex, Chugai Pharmaceutical and Nippon Boehringer Ingelheim. MI reports honoraria from Bayer, Bristol-Myers Squibb, Eli Lilly, Eisai, Sumitomo Dainippon Pharma, EA Pharma, Teijin Pharma, Yakult Honsha, Taiho Pharmaceutical, Otsuka, MSD, Mylan, Nihon Servier, Chugai Pharmaceutical, AstraZeneca, AbbVie, Abbott, Takeda, Novartis and Astellas Pharma; and advisory roles with Bayer, Eli Lilly, Eisai, Chugai Pharmaceutical, AstraZeneca, Takeda Pharmaceutical, Ono Pharmaceutical, GlaxoSmithKline and Nihon Servier; and grants from Bayer, Bristol-Myers Squibb, Eli Lilly, Eisai, Takeda Pharmaceutical, AstraZeneca, Chugai Pharmaceutical, Merck Biopharma, ASLAN Pharmaceuticals, Novartis, Yakult Honsha, Taiho Pharmaceutical, Ono Pharmaceutical, MSD, Merus N.V., Nihon Servier, Pfizer, Chiome Bioscience, Delta-Fly Pharma, and J-Pharma. CM reports honoraria from Nihon Servier, Novartis, Yakult Honsha, Teijin Pharma, Taiho Pharmaceutical, Eisai and MSD; advisory roles with Yakult Honsha, Novartis, Servier, Taiho Pharmaceutical and Abbvie; and grants from Eisai, Yakult Honsha, Ono Pharmaceutical, Taiho Pharmaceutical, J-Pharma, Daiichi Sankyo, HITACHI, AstraZeneca and Merck Biopharma.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and the Japanese Ethical Guidelines for Medical and Health Research Involving Human Subjects. The protocol of the GOZILA study was approved by the institutional review board of each participating institution and registered at the University Hospital Medical Information Network (UMIN) Clinical Trials Registry (protocol no. UMIN000029315 for GOZILA.
Consent to publish
This manuscript does not contain any individual person’s data such as individual details, images or videos.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Umemoto, K., Sunakawa, Y., Ueno, M. et al. Clinical significance of circulating-tumour DNA analysis by metastatic sites in pancreatic cancer. Br J Cancer 128, 1603–1608 (2023). https://doi.org/10.1038/s41416-023-02189-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02189-y
This article is cited by
-
High somatic mutations in circulating tumor DNA predict response of metastatic pancreatic ductal adenocarcinoma to first-line nab-paclitaxel plus S-1: prospective study
Journal of Translational Medicine (2024)
-
Current research status of tumor cell biomarker detection
Microsystems & Nanoengineering (2023)