Abstract
Background
Bronchial washing fluid (BWF) is a less-invasive specimen. Due to the limited sensitivity of BWF cellular component diagnosis, the aim of this study was to explore the potential role of BWF supernatant as a source of liquid biopsy of lung cancer.
Methods
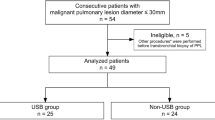
This prospective study enrolled 76 suspected and 5 progressed lung cancer patients. Transbronchial biopsy tissues, BWF supernatant (BWF_Sup) and BWF precipitant (BWF_Pre) were tested by a targeted panel of 1021 genes.
Results
BWF_Sup cell-free DNA (cfDNA) was superior to tissue biopsy and BWF_Pre in determining mutational allele frequency, tumour mutational burden, and chromosomal instability. Moreover, BWF_Sup and BWF_Pre achieved comparable efficacy to tissue samples in differentiating malignant and benign patients, but only BWF_Sup persisted differentiated performance after excluding 55 malignancies pathologically diagnosed by bronchoscopic biopsy. Among 67 malignant patients, 82.1% and 71.6% of tumour-derived mutations (TDMs) were detected in BWF_Sup and BWF_Pre, respectively, and the detectability of TDMs in BWF_Sup was independent of the cytological examination of BWF. BWF_Sup outperformed BWF_Pre in providing more subclonal information and thus might yield advantage in tracking drug-resistant markers.
Conclusions
BWF_Sup cfDNA is a reliable medium for lung cancer diagnosis and genomic profiles and may provide important information for subsequent therapeutic regimens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The datasets supporting the conclusions of this article are available from the corresponding author on reasonable request.
References
Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39.
Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6.
Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–46.
Lee CK, Davies L, Wu YL, Mitsudomi T, Inoue A, Rosell R, et al. Gefitinib or erlotinib vs chemotherapy for EGFR mutation-positive lung cancer: individual patient data meta-analysis of overall survival. J Natl Cancer Inst. 2017;109:djw279.
Lim C, Tsao MS, Le LW, Shepherd FA, Feld R, Burkes RL, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Annal Oncol. 2015;26:1415–21.
Aucamp J, Bronkhorst AJ, Badenhorst CPS, Pretorius PJ. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol Rev Camb Philos Soc. 2018;93:1649–83.
Pessoa LS, Heringer M, Ferrer VP. ctDNA as a cancer biomarker: a broad overview. Crit Rev Oncol/Hematol. 2020;155:103109.
Schrock AB, Welsh A, Chung JH, Pavlick D, Bernicker EH, Creelan BC, et al. Hybrid capture-based genomic profiling of circulating tumor DNA from patients with advanced non-small cell lung cancer. J Thorac Oncol. 2019;14:255–64.
Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–54.
Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. 2018;36:1631–41.
Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90.
Thompson JC, Yee SS, Troxel AB, Savitch SL, Fan R, Balli D, et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res. 2016;22:5772–82.
Reckamp KL, Melnikova VO, Karlovich C, Sequist LV, Camidge DR, Wakelee H, et al. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol. 2016;11:1690–700.
De Mattos-Arruda L, Mayor R, Ng CKY, Weigelt B, Martínez-Ricarte F, Torrejon D, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839.
Shin S, Kim J, Kim Y, Cho SM, Lee KA. Assessment of real-time PCR method for detection of EGFR mutation using both supernatant and cell pellet of malignant pleural effusion samples from non-small-cell lung cancer patients. Clin Chem Lab Med. 2017;55:1962–9.
Li YS, Jiang BY, Yang JJ, Zhang XC, Zhang Z, Ye JY, et al. Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of EGFR-mutant non-small-cell lung cancer: a new medium of liquid biopsy. Annal Oncol. 2018;29:945–52.
Zhai J, Han S, Guo Q, Shan B, Wang J, Guo Y, et al. Identifying genomic alterations in small cell lung cancer using the liquid biopsy of bronchial washing fluid. Front Oncol. 2021;11:647216.
Ryu JS, Lim JH, Lee MK, Lee SJ, Kim HJ, Kim MJ, et al. Feasibility of bronchial washing fluid-based approach to early-stage lung cancer diagnosis. Oncologist. 2019;24:e603–e6.
Kimura H, Kasahara K, Kawaishi M, Kunitoh H, Tamura T, Holloway B, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006;12:3915–21.
Nahar R, Zhai W, Zhang T, Takano A, Khng AJ, Lee YY, et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun. 2018;9:216.
Parikh AR, Leshchiner I, Elagina L, Goyal L, Levovitz C, Siravegna G, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med. 2019;25:1415–21.
Lin G, Li C, Li PS, Fang WZ, Xu HP, Gong YH, et al. Genomic origin and EGFR-TKI treatments of pulmonary adenosquamous carcinoma. Ann Oncol. 2020;31:517–24.
Chabon JJ, Hamilton EG, Kurtz DM, Esfahani MS, Moding EJ, Stehr H, et al. Integrating genomic features for non-invasive early lung cancer detection. Nature. 2020;580:245–51.
Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9.
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9.
Li J, Lupat R, Amarasinghe KC, Thompson ER, Doyle MA, Ryland GL, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics. 2012;28:1307–13.
Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49:211–5.
Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11:396–8.
Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–37.e10
Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376:2415–26.
Miller RJ, Casal RF, Lazarus DR, Ost DE, Eapen GA. Flexible bronchoscopy. Clin chest Med. 2018;39:1–16.
Zhang SJ, Zhang M, Zhou J, Zhang QD, Xu QQ, Xu X. Comparison of radial endobronchial ultrasound with a guide sheath and with distance by thin bronchoscopy for the diagnosis of peripheral pulmonary lesions: a prospective randomized crossover trial. J Thorac Dis. 2016;8:3112–8.
Schmidt B, Carstensen T, Engel E, Jandrig B, Witt C, Fleischhacker M. Detection of cell-free nucleic acids in bronchial lavage fluid supernatants from patients with lung cancer. Eur J Cancer. 2004;40:452–60.
Carstensen T, Schmidt B, Engel E, Jandrig B, Witt C, Fleischhacker M. Detection of cell-free DNA in bronchial lavage fluid supernatants of patients with lung cancer. Ann N. Y Acad Sci. 2004;1022:202–10.
Cardarella S, Ortiz TM, Joshi VA, Butaney M, Jackman DM, Kwiatkowski DJ, et al. The introduction of systematic genomic testing for patients with non-small-cell lung cancer. J Thorac Oncol. 2012;7:1767–74.
Herth FJ, Ernst A, Becker HD. Endobronchial ultrasound-guided transbronchial lung biopsy in solitary pulmonary nodules and peripheral lesions. Eur Respiratory J. 2002;20:972–4.
Guan Y, Hu H, Peng Y, Gong Y, Yi Y, Shao L, et al. Detection of inherited mutations for hereditary cancer using target enrichment and next generation sequencing. Fam Cancer. 2015;14:9–18.
Qu S, Chen Q, Yi Y, Shao K, Zhang W, Wang Y, et al. A reference system for BRCA mutation detection based on next-generation sequencing in the Chinese population. J Mol Diagn. 2019;21:677–86.
Zhang Y, Yao Y, Xu Y, Li L, Gong Y, Zhang K, et al. Pan-cancer circulating tumor DNA detection in over 10,000 Chinese patients. Nat Commun. 2021;12:11.
Liu X, Wang F, Xu C, Chen X, Hou X, Li Q, et al. Genomic origin and intratumor heterogeneity revealed by sequencing on carcinomatous and sarcomatous components of pulmonary sarcomatoid carcinoma. Oncogene. 2021;40:821–32.
Hu C, Zhao L, Liu W, Fan S, Liu J, Liu Y, et al. Genomic profiles and their associations with TMB, PD-L1 expression, and immune cell infiltration landscapes in synchronous multiple primary lung cancers. J Immunother Cancer. 2021;9:e003773.
Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16:398–406.
Pérez-Callejo D, Romero A, Provencio M, Torrente M. Liquid biopsy based biomarkers in non-small cell lung cancer for diagnosis and treatment monitoring. Transl Lung Cancer Res. 2016;5:455–65.
Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479–91.
Zhong WZ, Wang Q, Mao WM, Xu ST, Wu L, Shen Y, et al. Gefitinib versus vinorelbine plus cisplatin as adjuvant treatment for stage II-IIIA (N1-N2) EGFR-mutant NSCLC (ADJUVANT/CTONG1104): a randomised, open-label, phase 3 study. Lancet Oncol. 2018;19:139–48.
Harris FR, Kovtun IV, Smadbeck J, Multinu F, Jatoi A, Kosari F, et al. Quantification of somatic chromosomal rearrangements in circulating cell-free DNA from ovarian cancers. Sci Rep. 2016;6:29831.
Dong X, Fernandez-Salas E, Li E, Wang S. Elucidation of resistance mechanisms to second-generation ALK inhibitors alectinib and ceritinib in non-small cell lung cancer cells. Neoplasia. 2016;18:162–71.
Zhang X, Li C, Ye M, Hu Q, Hu J, Gong Z, et al. Bronchial washing fluid versus plasma and bronchoscopy biopsy samples for detecting epidermal growth factor receptor mutation status in lung cancer. Front Oncol. 2021;11:602402.
Zhou C, Yuan Z, Ma W, Qi L, Mahavongtrakul A, Li Y, et al. Clinical utility of tumor genomic profiling in patients with high plasma circulating tumor DNA burden or metabolically active tumors. J Hematol Oncol. 2018;11:129.
Zheng D, Ye X, Zhang MZ, Sun Y, Wang JY, Ni J, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep. 2016;6:20913.
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24.
Tong L, Ding N, Tong X, Li J, Zhang Y, Wang X, et al. Tumor-derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics. 2019;9:5532–41.
Acknowledgements
Prior abstract presentation: Oral presentation, Tumour-derived cell-free DNA from bronchoalveolar lavage fluid (BALF): a potential liquid biopsy analysis in lung cancer patients; April 3, 2019, American Association for Cancer Research (AACR) Annual Meeting, Atlanta, USA.
Funding
None.
Author information
Authors and Affiliations
Contributions
Prof. Xin Zhang is the guarantor of the content of the manuscript, including the data and analysis. Xinyu Zhang and YX had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. ZY, Yencheng Chao, QH, CL, MY, X Zhu, LC, JB, Y Gong, Y Guan, MZ, JH, HZ, TR, QS, KW, YH, XX, DPC and XP contributed substantially to the study design, data analysis and interpretation, and the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Zhongshan Hospital of Fudan University (No. B2018-027) and complied with all relevant ethical regulations. All participating patients signed written informed consent.
Consent to publish
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Yu, Z., Xu, Y. et al. Utility of cell-free DNA from bronchial washing fluid in diagnosis and genomic determination for radiology-suspected pulmonary nodules. Br J Cancer 127, 2154–2165 (2022). https://doi.org/10.1038/s41416-022-01969-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-022-01969-2