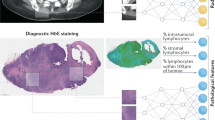
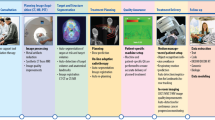
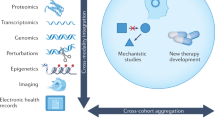
Abstract
The role of Artificial Intelligence and Machine Learning in cancer research offers several advantages, primarily scaling up the information processing and increasing the accuracy of the clinical decision-making. The key enabling tools currently in use in Precision, Digital and Translational Medicine, here named as ‘Intelligent Systems’ (IS), leverage unprecedented data volumes and aim to model their underlying heterogeneous influences and variables correlated with patients’ outcomes. As functionality and performance of IS are associated with complex diagnosis and therapy decisions, a rich spectrum of patterns and features detected in high-dimensional data may be critical for inference purposes. Many challenges are also present in such discovery task. First, the generation of interpretable model results from a mix of structured and unstructured input information. Second, the design, and implementation of automated clinical decision processes for drawing disease trajectories and patient profiles. Ultimately, the clinical impacts depend on the data effectively subjected to steps such as harmonisation, integration, validation, etc. The aim of this work is to discuss the transformative value of IS applied to multimodal data acquired through various interrelated cancer domains (high-throughput genomics, experimental biology, medical image processing, radiomics, patient electronic records, etc.).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
McNutt TR, Benedict SH, Low DA, Moore K, Shpitser I, Jiang W, et al. Using big data analytics to advance precision radiation oncology. Int J Radiat Oncol Biol Phys. 2018;101:285–91.
Yu B. Three principles of data science: predictability, computability, and stability. KDD ‘17: In: Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM Digital Library; 2017. p. 5.
Hulsen T, Jamuar SS, Moody AR, Karnes JH, Varga O, Hedested S, et al. From big data to precision medicine. Front Med. 2019;6:34.
Cheng F, Desai RJ, Handy DE, Wang R, Schneeweiss S, Barabasi A-L, et al. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat Commun. 2018;9:2691.
Kong J, Lee H, Kim D, Han SK, Ha D, Shin K, et al. Network-based machine learning in colorectal and bladder organoid models predicts anti-cancer drug efficacy in patients. Nat Commun. 2020;11:5485.
Kamdar MR, Fernández JD, Polleres A, Tudorache T, Musen MA. Enabling Web-scale data integration in biomedicine through linked open data. NPJ Digit Med. 2019;2:90.
Chan HCS, Shan H, Dahoun T, Vogel H, Yuan S. Advancing drug discovery via artificial intelligence. Trends Pharmacol Sci. 2019;40:592–604. Erratum in: Trends Pharmacol Sci. 2019;40:801.
Seyhan AA, Carini C. Are innovation and new technologies in precision medicine paving a new era in patients centric care? J Transl Med. 2019;17:114.
Koelzer VH, Sirinukunwattana K, Rittscher J, Mertz KD. Precision immunoprofiling by image analysis and artificial intelligence. Virchows Arch. 2019;474:511–22.
Leiserson MDM, Syrgkanis V, Gilson A, Dudik M, Gillett S, Chayes J, et al. A multifactorial model of T cell expansion and durable clinical benefit in response to a PD-L1 inhibitor. PLoS ONE. 2018;13:e0208422.
Snyder A, Nathanson T, Funt SA, Ahuja A, Buros Novik J, Hellmann MD, et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: An exploratory multi-omic analysis. PLoS Med. 2017;14:e1002309.
Parikh RB, Gdowski A, PAtt DA, Hertler A, Mermel C, Bekelman JE. Using big data and predictive analytics to determine patient risk in oncology. Am Soc Clin Oncol Educ Book. 2019;39:e53–e58.
Sechopoulos I, Mann RM. Stand-alone artificial intelligence—the future of breast cancer screening? Breast. 2020;49:254–60.
Kann BH, Thompson R, Thomas CR, Dicker A, Aneja S. Artificial intelligence in oncology: current applications and future directions. Oncology. 2019;33:45–63.
Patel SK, George B, Rai V. Artificial Intelligence to decode cancer mechanism: beyond patient stratification for precision oncology. Front Phys. 2020;11:1177.
Rattan R, Kataria T, Banerjee S, Goyal S, Gupta D, Pandita A, et al. Artificial intelligence in oncology, its scope and future prospects with specific reference to radiation oncology. Br J Radiol. 2019;1:1.
Weikert T, Cyriac J, Yang S, Nesic I, Parmar V, Stieltjes B. A practical guide to artificial intelligence based analysis in radiology. Invest Radiol. 2020;55:1–7.
Nagy M, Radakovich N, Nazha A. Machine learning in oncology: what should clinicians know? JCO Clin Cancer Inform. 2020;4:799–810.
Tseng H-H, Wei L, Luo Y, Ten Haken RK, El Naqa I. Machine learning and imaging informatics in oncology. Oncology. 2020;98:344–62.
Jaffray DA, Das S, Jacobs PM, Jeraj R, Lambin P. How advances in imaging will affect precision radiation oncology. Int J Radiat Oncol Biol Phys. 2018;101:292–8.
Esteva A, Kuprel B, Novoa R, Ko J, Swetteret SM, Blau HM. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–8.
Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. J Am Med Assoc. 2017;318:2199–210.
Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys. 2017;44:5162–71.
Levine AB, Schlosser C, Grewal J, Coope R, Jones SJM, Yip S. Rise of the machines: advances in deep learning for cancer diagnosis. Trends Canc. 2019;5:157–69.
Zhou J, Theesfeld CL, Yao K, Chen KM, Wong AK, Troyanskaya OG. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat Genet. 2918;50:1171–9.
Lambin P, Leijenaar RTH, Deist TM, Perlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev. 2017;14:749–62.
Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–10.
Thrall JH, Li X, Li Q, Cruz C, Do S, Dreyer K, et al. Artificial intelligence and machine learning in radiology: opportunities, challeneges, pitfalls, and criteria for success. J Am Coll Radiol. 2018;15:504–8.
Parekh VS, Jacobs MA. Deep learning and radiomics in precision medicine. Exp Rev Precis Med Drug Dev. 2019;4:59–72.
Avanzo M, Wei L, Stancanello J, Vallières M, Rao A, Morin O, et al. Machine and deep learning methods for radiomics. Med Phys. 2020;47:e185–202.
Sakellaropoulos T, Vougas K, Narang S, Koinis F, Kotsinas A, et al. A deep learning framework for predicting response to therapy in cancer. Cell Rep. 2019;29:3367–73.
Liang G, Fan W, Luo H, Zhu X. The emerging roles of artificial intelligence in cancer drug development and precision therapy. Biomed Pharmacother. 2020;128:110255.
Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22:976–86.
Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–48.e24.
Segler MHS, Preuss M, Waller MP. Planning chemical syntheses with deep neural networks and symbolic AI. Nature. 2018;555:604.
Vamathevan J, Clark D, Czodrowski P, Dunham I, Ferran E, Lee G, et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18:463–77.
Dinić J, Efferth T, García-Sosa AT, Grahovac J, Padrón JM, Pajeva I, et al. Repurposing old drugs to fight multidrug resistant cancers. Drug Resist Updat. 2020;52:100713.
Bulik-Sullivan B, Busby J, Palmer CD, Davis MJ, Murphy T, Clark A, et al. Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat Biotechnol. 2019;37:55–63.
Nazha A, Sekeres MA, Bejar R, Rauh MJ, Othus M, Komrokji RS, et al. Genomic biomarkers to predict resistance to hypomethylating agents in patients with myelodysplastic syndromes using artificial intelligence. JCO Prec Oncol. 2019;3:1–11.
Nasief H, Zheng C, Schott D, Hall W, Tsai S, Erickson B, et al. A machine learning based delta-radiomics process for early prediction of treatment response of pancreatic cancer. npj Precis Oncol. 2019;3:25.
Lou B, Doken S, Zhuang T, Wingerter D, Gidwani M, Mistry N, et al. An image-based deep learning framework for individualizing radiotherapy dose. Lancet Digit Health. 2019;1:e136–147. Erratum in: Lancet Digit Health. 2019;1:e160.
Hou Z, Ren W, Li S, Liu J, Sun Y, Yan J, et al. Radiomic analysis in contrast-enhanced CT: predict treatment response to chemoradiotherapy in esophageal carcinoma. Oncotarget. 2017;8:104444–54.
Nguyen D, Long T, Jia X, Lu W, Gu X, Iqbal Z, et al. A feasibility study for predicting optimal radiation therapy dose distributions of prostate cancer patients from patient anatomy using deep learning. Sci Rep. 2019;9:1076.
Nguyen D, Jia X, Sher D, Lin MH, Iqbal Z, Liu H, et al. D radiotherapy dose prediction on head and neck cancer patients with a hierarchically densely connected U-net deep learning architecture. Phys Med Biol. 2019;64:065020.
Hollon TC, Pandian B, Adapa AR, Urias E, Save AV, Khalsa SSS, et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med. 2020;26:52–58.
Halabi S, Li C, Luo S. Developing and validating risk assessment models of clinical outcomes in modern oncology. JCO Prec Oncol. 2019;3:PO.19.000068.
Blyuss O, Zaikin A, Cherepanova V, Munblit D, Kiseleva EM, Prytomanova OG, et al. Development of PancRISK, a urine biomarker-based risk score for stratified screening of pancreatic cancer patients. Br J Cancer. 2020;122:692–6.
Kim D, Joung J-G, Sohn K-A, Shin H, Park YR, Ritchie MD, et al. Knowledge boosting: a graph-based integration approach with multi-omics data and genomic knowledge for cancer clinical outcome prediction. J Am Med Inform Assoc. 2015;22:109–20.
Cook DP, Vanderhyden BC. Context specificity of the EMT transcriptional response. Nat Commun. 2020;11:2142.
Lipinski KA, Barber LJ, Davies MN, Ashenden M, Sottoriva A, Gerlinger M. Cancer evolution and the limits of predictability in precision cancer medicine. Trends Cancer. 2016;2:49–63.
Azuaje F. Artificial Intelligence for precision oncology: beyond patient stratification. Npj Prec Oncol. 2019;3:6.
Pan SJ, Yang Q. A survey on transfer learning. IEEE Tr Knowl Data Eng. 2010;22:1345–59.
Turki T, Wei Z, Wang, TL J. A transfer learning approach via procrustes analysis and mean shift for cancer drug sensitivity prediction. J Bioinform Comput Biol. 2018;16:1840014.
Sevakula RK, Singh V, Verma NK, Kumar C, Cui Y. Transfer learning for molecular cancer classification using deep neural networks. IEEE/ACM Trans Comput Biol Bioinform. 2019;16:2089–2100.
Vu CC, Siddiqui ZA, Zamdborg L, Thompson AB, Quinn TJ, Castillo E, et al. Deep convolutional neural networks for automatic segmentation of thoracic organs-at-risk in radiation oncology - use of non-domain transfer learning. J Appl Clin Med Phys. 2020;21:108–13.
Poudel P, Nyamundanda G, Patil Y, Cheang MCU, Sadanandam A. Heterocellular gene signatures reveal luminal-A breast cancer heterogeneity and differential therapeutic responses. npj Breast Cancer. 2019;5:21.
Kather JN, Heij LR, Grabsch HI, Loeffler C, Echle A, Muti HS, et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer. 2020;1:789–99.
Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Canc J Clin. 2019;69:127–57.
Blank CU, Haanen JB, Ribas A, Schumacher TN. Cancer Immunology. The “cancer immunogram”. Science. 2016;352:658–60.
Lyons YA, Wu SY, Overwijk WW, Baggerly KA, Sood AK. Immune cell profiling in cancer: molecular approaches to cell-specific identification. npj Prec Oncol. 2017;1:26.
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority oversampling technique. J Art Intell Res. 2002;16:321–257.
He H, Bai Y, Garcia EA, Li S. ADASYN: adaptive synthetic sampling approach for imbalanced learning. In: 2008 IEEE international joint conference on neural networks. (IEEE Xplore ed.), IEEE; 2008. p. 1322–8.
Griffith GJ, Morris TT, Tudball MJ, Herbert A, Mancano G, Pike L, et al. Collider bias undermines our understanding of COVID-19 disease risk and severity. Nat Commun. 2020;11:5749.
Bueno MJ, Mouron S, Quintela-Fandino M. Personalising and targeting antiangiogenic resistance: a complex and multifactorial approach. Br J Cancer. 2017;116:1119–25.
Dlamini Z, Francies FZ, Hull R, Marima R. Artificial intelligence (AI) and big data in cancer and precision oncology. Computat Str Biotech J. 2020;18:2300–11.
Halama N. Machine learning for tissue diagnostics in oncology: brave new world. Br J Cancer. 2019;121:431–3.
Tuffaha HW, Gordon LG, Scuffham PA. Value of information analysis in oncology: the value of evidence and evidence of value. J Oncol Pract. 2014;10:e55–62.
Kunst NR, Alarid-Escudero F, Paltiel AD, Wang S-Y. A value of information analysis of research on the 21-gene assay for breast cancer management. Value Health. 2019;22:1102–10.
Beaton L, Bandula S, Gaze MN, Sharma RA. How rapid advances in imaging are defining the future of precision radiation oncology. Br J Cancer. 2019;120:779–90.
Linn KA, Laber EB, Stefanski LA. iqLearn: interactive Q-Learning in R. J Stat Softw. 2015;64:i01.
Tseng HH, Luo Y, Cui S, Chien JT, Ten Haken RK, El Naqa I. Deep reinforcement learning for automated radiation adaptation in lung cancer. Med Phys. 2017;44:6690–705.
Petersen BK, Yang J, Grathwohl WS, Cockrell C, Santiago C, An G, et al. Deep reinforcement learning and simulation as a path toward precision medicine. J Comput Biol. 2019;26:597–604.
Ali I, Hart GR, Gunabushanam G, Liang Y, Muhammad W, Nartowt B, et al. Lung nodule detection via deep reinforcement learning. Front Oncol. 2018;8:108.
Liu S, See KC, Ngiam KY, Celi LA, Feng M. Reinforcement learning for clinical decision support in critical care: comprehensive review. J Med Intern Res. 2020;2287:e18477.
Mazurowski MA. Radiogenomics: what it is and why it is important. J Am Coll Radiol. 2015;12:862–6.
Wu J, Tha KK, Xing L, Li R. Radiomics and radiogenomics for precision radiotherapy. J Radiat Res. 2018;59:i25–i31.
Papanikolaou N, Matos C, Koh DM. How to develop a meaningful radiomic signature for clinical use in oncologic patients. Cancer Imag. 2020;20:33.
Keek SA, Leijenaar RTH, Jochems A, Woodruff HC. A review on radiomics and the future of theranostics for patient selection in precision medicine. Br J Radiol. 2018;91:20170926.
Alvarez-Jimenez C, Sandino AA, Prasanna P, Gupta A, Viswanath SE, Romero E. Identifying cross-scale associations between radiomic and pathomic signatures of non-small cell lung cancer subtypes: preliminary results. Cancers. 2020;12:3663.
Saltz JH, Gupta R. Artificial intelligence and the interplay between tumor and immunity, Ch. 10. In: Artificial Intelligence and Deep Learning in Pathology. (Stanley C ed.), Elsevier; 2021. p. 211–35.
Nie K, Al-Hallaq H, Li A, Benedict SH, Sohn JW, Moran JM, et al. NCTN assessment of current applications of radiomics in oncology. Int J Rad Oncol. 2019;104:302–15.
Lv W, Ashrafinia S, Ma J, Lu L, Rahmim A. Multi-level multi-modality fusion radiomics: application to PET and CT imaging for prognostication of head and neck. Cancer IEEE J Biomed Health Inform. 2020;24:2268–77.
Wei L, Osman S, Hatt M, El Naqa I. Machine learning for radiomics-based multimodality and multiparametric modeling. Q J Nucl Med Mol Imag. 2019;63:323–38.
Papp L, Spielvogel CP, Rausch I, Hacker M, Beyer T. Personalized medicine through hybrid imaging and medical big data analysis. Front Phys. 2018;6:51.
Hagiwara A, Fujita S, Ohno M, Aoki S. Variability and standardization of quantitative imaging. Integr Radiol. 2020;55:601–16.
Mühlberg A, Katzmann A, Heinemann V, Kärgel R, Wels M, Taubmann O, et al. The Technome—a predictive internal calibration approach for quantitative imaging biomarker research. Sci Rep. 2020;10:1103.
Sala E, Merna E, Himoto Y, Veeraraghavan H, Brenton JD, Snyder A, et al. Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol. 2017;72:3–10.
Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are. Data Radiol. 2016;278:563–77.
Gillies RJ, Balagurunathan Y. Perfusion MR imaging of breast cancer: insights using ‘habitat imaging’. Radiology. 2018;288:36–37.
Sollini M, Antunovic L, Chiti A, Kirienko M. Towards clinical application of image mining: a systematic review on artificial intelligence and radiomics. Eur J Nucl Med Mol Imag. 2019;46:2656–72.
Fave X, Zhang L, Yang J, Mackin D, Balter P, Gomez D, et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep. 2017;7:588.
Jeon SH, Song C, Chie EK, Kim B, Kim YH, Chang W, et al. Delta-radiomics signature predicts treatment outcomes after preoperative chemoradiotherapy and surgery in rectal cancer. Radiat Oncol. 2019;14:43.
Lin P, Yang PF, Chen S, Shao Y-Y, Xu L, Wu Y, et al. A delta-radiomics model for preoperative evaluation of neoadjuvant chemotherapy response in high-grade osteosarcoma. Cancer Imag. 2020;20:7.
Gatouillat A, Badr Y, Massot B, Sejdić E. Internet of medical things: a review of recent contributions dealing with cyber-physical systems in medicine. IEEE Internet Things J. 2018;5:3810–22.
Han T, Nunes VX, Souza LFDF, Marques AG, Silva ICL, Marcos Aurelio AF, et al. Internet of medical things—based on deep learning techniques for segmentation of lung and stroke regions in CT scans. IEEE Access. 2020;8:71117–35.
Souza LFF, Silva ICL, Marques AG, Silva FHDS, Nunes VX, Hassan MM, et al. Internet of medical things: an effective and fully automatic iot approach using deep learning and fine-tuning to lung CT segmentation. Sensors. 2020;20:E6711.
Sun C, Tian X, Liu Z, Li W, Li P, Chen J, et al. Radiomic analysis for pretreatment prediction of response to neoadjuvant chemotherapy in locally advanced cervical cancer: a multicentre study. eBioMedicine. 2019;46:160–9.
Dissaux G, Visvikis D, Da-Ano R, Pradier O, Chajon E, Barillot I, et al. Pretreatment 18F-FDG PET/CT radiomics predict local recurrence in patients treated with stereotactic body radiotherapy for early-stage non-small cell lung cancer: a multicentric study. J Nucl Med. 2020;61:814–20.
Li ZC, Bai H, Sun Q, Li Q, Liu L, Zou Y, et al. Multiregional radiomics features from multiparametric MRI for prediction of MGMT methylation status in glioblastoma multiforme: a multicentre study. Eur Radiol. 2018;28:3640–50.
Welch ML, McIntosh C, Haibe-Kains B, Milosevic MF, Wee L, Dekker A, et al. Vulnerabilities of radiomic signature development: the need for safeguards. Radioth Oncol. 2019;130:2–9.
Capobianco E, Valdes C, Sarti S, Jiang Z, Poliseno L, Tsinoremas NF. Ensemble modeling approach targeting heterogeneous RNA-Seq data: application to melanoma pseudogenes. Sci Rep. 2017;7:17344.
Ho D. Artificial intelligence in cancer therapy. Science. 2020;367:982–3.
Shah P, Kendall F, Khozin S, Goosen R, Hu J, Laramie J, et al. Artificial intelligence and machine learning in clinical development: a translational perspective. npj Digit Med. 2019;2:69.
Toh TS, Dondelinger F, Wang D. Looking beyond the hype: applied AI and machine learning in translational medicine. EBioMedicine 2019;47:607–15.
Liu R, Rizzo S, Whipple S, Pal N, Pineda AL, Lu M, et al. Evaluating eligibility criteria of oncology trials using real-world data and AI. Nature 2021.
Capobianco E. Imprecise data and their impact on translational research in medicine. Front Med. 2020;7:82.
Bezemer T, de Groot MC, Blasse E, Ten Berg MJ, Kappen TH, Bredenoord AL, et al. Factor in clinical decision support systems. J Med Intern Res. 2019;21:e11732.
Luo H, Zhao Q, Wei W, Zheng L, Yi S, Li G, et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med. 2020;12:eaax7533. Erratum in: Sci Transl Med. 2020;12:eabc1078.
Liu Y, Kohlberger T, Norouzi M, Dahl GE, Smith JL, Mohtashamian A, et al. Artificial intelligence–based breast cancer nodal metastasis detection: insights into the black box for pathologists. Arch Pathol Lab Med. 2019;143:859–68.
Steiner DF, MacDonald R, Liu Y, Truszkowski P, Hipp JD, Gammage C, et al. Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am J Surg Path. 2018;42:1636–46.
Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019;17:195.
Collins GS, Moons KGM. Reporting of artificial intelligence prediction models. Lancet. 2019;393:1577–9.
Faes L, Liu X, Wagner SK, Fu DJ, Balaskas KA. Clinician’s guide to artificial intelligence: how to critically appraise machine learning studies. Transl Vis Sci Technol. 2020;9:33. Erratum in: Transl Vis Sci Technol. 2020;9:7.
CONSORT-AI and SPIRIT-AI Steering Group. Reporting guidelines for clinical trials evaluating artificial intelligence interventions are needed. Nat Med. 2019;25:1467–8.
Liu X, Faes L, Calvert MJ, Denniston AK. CONSORT/SPIRIT-AI Extension Group. Extension of the CONSORT and SPIRIT statements. Lancet. 2019;394:1225.
Dong Y, Yang W, Wang J, Zhao J, Qiang Y. MLW-gcForest: a multi-weighted gcForest model towards the staging of lung adenocarcinoma based on multi-modal genetic data. BMC Bioinform. 2019;20:578.
Nestor B, McDermott MBA, Chauhan G, Naumann T, Hughes MC, Goldenberg A, et al. Rethinking clinical prediction: why machine learning must consider year of care and feature aggregation. In: Machine Learning for Health (ML4H): Workshop at NeurIPS. 2018. arXiv:1811.07216 [cs.LG].
Davis SE, Greevy RA, Fonnesbeck C, Lasko TA, Walsh CG, Matheny ME. A nonparametric updating method to correct clinical prediction model drift. J Am Med Inform Assoc. 2019;26:1448–57.
Acknowledgements
The author acknowledges NSF support from grant NSF 19-500. DMS 1918925/1922843 (years: 08/01/2019 – 08/01/2022).
Funding
None.
Author information
Authors and Affiliations
Contributions
All contributions were from the single author.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Competing interests
The author declares no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Capobianco, E. High-dimensional role of AI and machine learning in cancer research. Br J Cancer 126, 523–532 (2022). https://doi.org/10.1038/s41416-021-01689-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01689-z
This article is cited by
-
Edge intelligence-assisted animation design with large models: a survey
Journal of Cloud Computing (2024)
-
Deep neural network based tissue deconvolution of circulating tumor cell RNA
Journal of Translational Medicine (2023)
-
UK Biobank: a globally important resource for cancer research
British Journal of Cancer (2023)