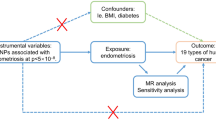
Abstract
Background
Genetic correlations, causalities and pathways between large-scale complex exposures and ovarian and breast cancers need systematic exploration.
Methods
Mendelian randomisation (MR) and genetic correlation (GC) were used to identify causal biomarkers from 95 cancer-related exposures for risk of breast cancer [BC: oestrogen receptor-positive (ER + BC) and oestrogen receptor-negative (ER − BC) subtypes] and ovarian cancer [OC: high-grade serous (HGSOC), low-grade serous, invasive mucinous (IMOC), endometrioid (EOC) and clear cell (CCOC) subtypes].
Results
Of 31 identified robust risk factors, 16 were new causal biomarkers for BC and OC. Body mass index (BMI), body fat mass (BFM), comparative body size at age 10 (CBS-10), waist circumference (WC) and education attainment were shared risk factors for overall BC and OC. Childhood obesity, BMI, CBS-10, WC, schizophrenia and age at menopause were significantly associated with ER + BC and ER − BC. Omega-6:omega-3 fatty acids, body fat-free mass and basal metabolic rate were positively associated with CCOC and EOC; BFM, linoleic acid, omega-6 fatty acids, CBS-10 and birth weight were significantly associated with IMOC; and body fat percentage, BFM and adiponectin were significantly associated with HGSOC. Both GC and MR identified 13 shared factors. Factors were stratified into five priority levels, and visual causal networks were constructed for future interventions.
Conclusions
With analysis of large-scale exposures for breast and ovarian cancers, causalities, genetic correlations, shared or specific factors, risk factor priority and causal pathways and networks were identified.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Researchers may have access to this data from the original researches shown in the supplement material.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ, et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J Clin. 2018;68:31–54.
Wilson LF, Antonsson A, Green AC, Jordan SJ, Kendall BJ, Nagle CM, et al. How many cancer cases and deaths are potentially preventable? Estimates for Australia in 2013. Int J Cancer. 2018;142:691–701.
Brown KF, Rumgay H, Dunlop C, Ryan M, Quartly F, Cox A, et al. The fraction of cancer attributable to modifiable risk factors in England, Wales, Scotland, Northern Ireland, and the United Kingdom in 2015. Br J Cancer. 2018;118:1130–41.
Zhu J, Jiang X, Niu Z. Alcohol consumption and risk of breast and ovarian cancer: a Mendelian randomization study. Cancer Genet. 2020;245:35–41.
Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41.
Beeghly-Fadiel A, Khankari NK, Delahanty RJ, Shu X-O, Lu Y, Schmidt MK, et al. A Mendelian randomization analysis of circulating lipid traits and breast cancer risk. Int J Epidemiol. 2020;49:1117–31.
Yarmolinsky J, Relton CL, Lophatananon A, Muir K, Menon U, Gentry-Maharaj A, et al. Appraising the role of previously reported risk factors in epithelial ovarian cancer risk: a Mendelian randomization analysis. PLoS Med. 2019;16:e1002893.
Au Yeung SL, Schooling CM. Impact of glycemic traits, type 2 diabetes and metformin use on breast and prostate cancer risk: a Mendelian randomization study. BMJ Open Diabetes Res Care. 2019;7:e000872.
Ooi BNS, Loh H, Ho PJ, Milne RL, Giles G, Gao C, et al. The genetic interplay between body mass index, breast size and breast cancer risk: a Mendelian randomization analysis. Int J Epidemiol. 2019;48:781–94.
Si S, Tewara MA, Li Y, Li W, Chen X, Yuan T, et al. Causal pathways from body components and regional fat to extensive metabolic phenotypes: a Mendelian randomization study. Obesity. 2020;28:1536–49.
Manning AK, Hivert M-F, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–69.
Bradfield JP, Taal HR, Timpson NJ, Scherag A, Lecoeur C, Warrington NM, et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44:526–31.
van der Valk RJP, Kreiner-Møller E, Kooijman MN, Guxens M, Stergiakouli E, Sääf A, et al. A novel common variant in DCST2 is associated with length in early life and height in adulthood. Hum Mol Genet. 2015;24:1155–68.
Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–44.
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7.
Soranzo N, Sanna S, Wheeler E, Gieger C, Radke D, Dupuis J, et al. Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–39.
Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–86.
Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83.
Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–21.
Lemaitre RN, Tanaka T, Tang W, Manichaikul A, Foy M, Kabagambe EK, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet. 2011;7:e1002193.
International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium 2, Sawcer S, Hellenthal G, Pirinen M, Spencer CCA, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–9.
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206.
Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45:145–54.
Felix JF, Bradfield JP, Monnereau C, van der Valk RJP, Stergiakouli E, Chesi A, et al. Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum Mol Genet. 2016;25:389–403.
Strawbridge RJ, Dupuis J, Prokopenko I, Barker A, Ahlqvist E, Rybin D, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–34.
Ahola-Olli AV, Würtz P, Havulinna AS, Aalto K, Pitkänen N, Lehtimäki T, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet. 2017;100:40–50.
Evans DM, Zhu G, Dy V, Heath AC, Madden PAF, Kemp JP, et al. Genome-wide association study identifies loci affecting blood copper, selenium and zinc. Hum Mol Genet. 2013;22:3998–4006.
Jiang X, O’Reilly PF, Aschard H, Hsu Y-H, Richards JB, Dupuis J, et al. Genome-wide association study in 79,366 European-ancestry individuals informs the genetic architecture of 25-hydroxyvitamin D levels. Nat Commun. 2018;9:260.
Clarke T-K, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, et al. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol Psychiatry. 2017;22:1376–84.
Klimentidis YC, Raichlen DA, Bea J, Garcia DO, Wineinger NE, Mandarino LJ, et al. Genome-wide association study of habitual physical activity in over 377,000 UK Biobank participants identifies multiple variants including CADM2 and APOE. Int J Obes (Lond). 2018;42:1161–76.
Kilpeläinen TO, Carli JFM, Skowronski AA, Sun Q, Kriebel J, Feitosa MF, et al. Genome-wide meta-analysis uncovers novel loci influencing circulating leptin levels. Nat Commun. 2016;7:10494.
Kettunen J, Demirkan A, Würtz P, Draisma HHM, Haller T, Rawal R, et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:11122.
DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, Mexican American Type 2 Diabetes (MAT2D) Consortium, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium, Mahajan A, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–44.
Meddens SFW, de Vlaming R, Bowers P, Burik CAP, Linnér RK, Lee C, et al. Genomic analysis of diet composition finds novel loci and associations with health and lifestyle. Mol Psychiatry. 2021;26:2056–69.
Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–9.
Doherty A, Smith-Byrne K, Ferreira T, Holmes MV, Holmes C, Pulit SL, et al. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat Commun. 2018;9:5257.
Day FR, Ruth KS, Thompson DJ, Lunetta KL, Pervjakova N, Chasman DI, et al. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. 2015;47:1294–303.
Warrington NM, Beaumont RN, Horikoshi M, Day FR, Helgeland Ø, Laurin C, et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat Genet. 2019;51:804–14.
Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WOC, Altmüller J, Ang W, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018;50:42–53.
Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16.
Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Mägi R, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–96.
Benyamin B, Esko T, Ried JS, Radhakrishnan A, Vermeulen SH, Traglia M, et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat Commun. 2014;5:4926.
Dastani Z, Hivert M-F, Timpson N, Perry JRB, Yuan X, Scott RA, et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8:e1002607.
Perry JR, Day F, Elks CE, Sulem P, Thompson DJ, Ferreira T, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–7.
Ferreira MA, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017;49:1752–7.
Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19.
Michailidou K, Lindström S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–4.
Phelan CM, Kuchenbaecker KB, Tyrer JP, Kar SP, Lawrenson K, Winham SJ, et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat Genet. 2017;49:680–91.
Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377–89.
Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35:1880–906.
Huang T, Afzal S, Yu C, Guo Y, Bian Z, Yang L, et al. Vitamin D and cause-specific vascular disease and mortality: a Mendelian randomisation study involving 99,012 Chinese and 106,911 European adults. BMC Med. 2019;17:160.
Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65.
Bowden J, Davey, Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14.
Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251–60.
Si S, Hou L, Chen X, Li W, Liu X, Liu C, et al. Exploring the causal roles of circulating remnant lipid profile on cardiovascular and cerebrovascular diseases: Mendelian randomization study. J Epidemiol. 2021 Jan 13. https://doi.org/10.2188/jea.JE20200305. Epub ahead of print. PMID: 33441507.
Si S, Li J, Tewara MA, Xue F. Genetically determined chronic low-grade inflammation and hundreds of health outcomes in the UK Biobank and the FinnGen population: a phenome-wide Mendelian randomization study. Front Immunol. 2021;12:720876.
Burgess S, Daniel RM, Butterworth AS, Thompson SG. EPIC-InterAct Consortium. Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int J Epidemiol. 2015;44:484–95.
Adams CD, Neuhausen SL. Bi-directional Mendelian randomization of epithelial ovarian cancer and schizophrenia and uni-directional Mendelian randomization of schizophrenia on circulating 1- or 2-glycerophosphocholine metabolites. Mol Genet Metab Rep. 2019;21:100539.
Dimou NL, Papadimitriou N, Mariosa D, Johansson M, Brennan P, Peters U, et al. Circulating adipokine concentrations and risk of five obesity-related cancers: a Mendelian randomization study. Int J Cancer. 2021;148:1625–36.
Yuan S, Xiong Y, Michaëlsson M, Michaëlsson K, Larsson SC. Genetically predicted education attainment in relation to somatic and mental health. Sci Rep. 2021;11:4296.
Qian F, Wang S, Mitchell J, McGuffog L, Barrowdale D, Leslie G, et al. Height and body mass index as modifiers of breast cancer risk in BRCA1/2 mutation carriers: a Mendelian randomization study. J Natl Cancer Inst. 2019;111:350–64.
Murphy N, Knuppel A, Papadimitriou N, Martin RM, Tsilidis KK, Smith-Byrne K, et al. Insulin-like growth factor-1, insulin-like growth factor-binding protein-3, and breast cancer risk: observational and Mendelian randomization analyses with ∼430 000 women. Ann Oncol. 2020;31:641–9.
Larsson SC, Carter P, Vithayathil M, Kar S, Mason AM, Burgess S. Insulin-like growth factor-1 and site-specific cancers: a Mendelian randomization study. Cancer Med. 2020;9:6836–42.
Richmond RC, Anderson EL, Dashti HS, Jones SE, Lane JM, Strand LB, et al. Investigating causal relations between sleep traits and risk of breast cancer in women: mendelian randomisation study. BMJ. 2019;365:l2327.
Byrne EM, Ferreira MAR, Xue A, Lindström S, Jiang X, Yang J, et al. Is schizophrenia a risk factor for breast cancer? Evidence from genetic data. Schizophr Bull. 2019;45:1251–6.
Robinson T, Martin RM, Yarmolinsky J. Mendelian randomisation analysis of circulating adipokines and C-reactive protein on breast cancer risk. Int J Cancer. 2020;147:1597–603.
Gao C, Patel CJ, Michailidou K, Peters U, Gong J, Schildkraut J, et al. Mendelian randomization study of adiposity-related traits and risk of breast, ovarian, prostate, lung and colorectal cancer. Int J Epidemiol. 2016;45:896–908.
Papadimitriou N, Dimou N, Tsilidis KK, Banbury B, Martin RM, Lewis SJ, et al. Physical activity and risks of breast and colorectal cancer: a Mendelian randomisation analysis. Nat Commun. 2020;11:597.
Dimou NL, Papadimitriou N, Gill D, Christakoudi S, Murphy N, Gunter MJ, et al. Sex hormone binding globulin and risk of breast cancer: a Mendelian randomization study. Int J Epidemiol. 2019;48:807–16.
Kar SP, Andrulis IL, Brenner H, Burgess S, Chang-Claude J, Considine D, et al. The association between weight at birth and breast cancer risk revisited using Mendelian randomisation. Eur J Epidemiol. 2019;34:591–600.
Richardson TG, Sanderson E, Elsworth B, Tilling K, Davey Smith G. Use of genetic variation to separate the effects of early and later life adiposity on disease risk: mendelian randomisation study. BMJ. 2020;369:m1203.
Saini RK, Keum Y-S. Omega-3 and omega-6 polyunsaturated fatty acids: dietary sources, metabolism, and significance—a review. Life Sci. 2018;203:255–67.
Adkins Y, Kelley DS. Mechanisms underlying the cardioprotective effects of omega-3 polyunsaturated fatty acids. J Nutr Biochem. 2010;21:781–92.
Zhou H, Zhang Y, Liu J, Yang Y, Fang W, Hong S, et al. Education and lung cancer: a Mendelian randomization study. Int J Epidemiol. 2019;48:743–50.
Day FR, Thompson DJ, Helgason H, Chasman DI, Finucane H, Sulem P, et al. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat Genet. 2017;49:834–41.
Qi G, Chatterjee N. Mendelian randomization analysis using mixture models for robust and efficient estimation of causal effects. Nat Commun. 2019;10:1941.
Acknowledgements
We thank to the MR Base platform, Integrative Epidemiology Unit (IEU) project, Neale Lab (Neale Lab analysis of UK Biobank phenotypes, round 2), MRC-IEU (MRC-IEU UK Biobank GWAS pipeline v 2), Borges et al. (metabolic biomarkers in the UK Biobank measured by Nightingale Health 2020), all related consortiums including BCAC, OCAC, GIANT, EGG, MRC-IEU, GUGC, Neale Lab, DIAGRAM, IMSGC, PGC, SSGAC, UK Biobank, GSCAN, GLGC, MAGIC, ADIPOGen, GIS, CHARGE and ReproGen, and original research by Astle et al., Jiang et al., Sun et al., Kettunen et al., Clarke et al., Doherty et al., Ferreira et al., Demenais et al., Klimentidis et al., Strawbridge et al., Manning et al., Dupuis et al., Evans et al. and Borges et al. for providing open access summary-level data resources. For a full description of acknowledgements concerning these studies, cohorts and consortiums, as well as funding information and other details, see Supplementary Text 3. We also thank the Charlesworth Group (https://www.cwauthors.com.cn/) for its linguistic assistance during the round 2 revision.
Funding
This research was funded by the National Key Research and Development Program (2020YFC2003500), the National Natural Science Foundation of China (81773547) and the Natural Science Foundation of Shandong Province (ZR2019ZD02). The corresponding author FX obtained the funding. The funders had no role in this work.
Author information
Authors and Affiliations
Contributions
SS and FX have the conception. SS did the statistical analyses and drafted the initial manuscript. MAT completed the revision of English grammar. All authors participated in the interpretation of the results, edited and reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Our research is only based on publicly available summarised data and no individual data were involved. These genome-wide association studies (including the GIANT, EGG, MRC-IEU, GUGC, Neale Lab, DIAGRAM, IMSGC, PGC, SSGAC, UK Biobank, GSCAN, GLGC, MAGIC, ADIPOGen, GIS, CHARGE and ReproGen consortium, and the individual research by Astle WJ et al, Jiang X et al, Sun BB et al, Kettunen et al, T-K Clarke et al, Aiden Doherty et al, Ferreira MA et al, Demenais F et al, Klimentidis YC et al, Strawbridge RJ et al, Manning AK et al, Dupuis J et al, Evans et al and Borges CM et al) had declared the ethic approval from the relevant institutional review board and in accordance with the declaration of Helsinki, or other (see reference [15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50]). This secondary analysis is based on summary-level statistics that are not applicable for the clauses of ethics, the Ethics Committee of the School of Public Health of Shandong University ruled that ethics approval was not required in this particular case.
Consent for publication
Not applicable
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Si, S., Li, J., Tewara, M.A. et al. Identifying causality, genetic correlation, priority and pathways of large-scale complex exposures of breast and ovarian cancers. Br J Cancer 125, 1570–1581 (2021). https://doi.org/10.1038/s41416-021-01576-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01576-7
This article is cited by
-
Relationship between oily fish intake and breast cancer based on estrogen receptor status: a Mendelian randomization study
Breast Cancer Research and Treatment (2024)
-
Ovarian cancer risk of Chinese women with BRCA1/2 germline pathogenic variants
Journal of Human Genetics (2022)