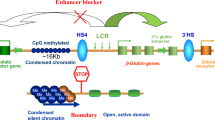
Abstract
Lymphoid-specific helicase (LSH) is a member of the SNF2 helicase family of chromatin-remodelling proteins. Dysfunctions or mutations in LSH causes an autosomal recessive disease known as immunodeficiency-centromeric instability-facial anomaly (ICF) syndrome. Interestingly, LSH participates in various aspects of epigenetic regulation, including nucleosome remodelling, DNA methylation, histone modifications and heterochromatin formation. Further, LSH plays a crucial role during DNA-damage repair, specifically during double-strand break (DSB) repair, since murine LSH was shown to be essential for non-homologous end joining (NHEJ) and homologous recombination (HR). Accordingly, overexpression of LSH drives tumorigenesis and malignancy. On the other hand, LSH homologs stabilise the genome. Thus, LSH might be implemented as a biomarker for various cancer types and potential target molecule to develop therapeutic strategies against them. In this review, we focus on the role of LSH in orchestrating chromatin rearrangements, such as DNA methylation and histone modifications, as well as in DNA-damage repair. Changes in chromatin structure may facilitate gene expression signatures that cause malignant transformation. We summarise recent findings of LSH in cancers and raise critical open questions for further studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data analysed in Fig. 2 were extracted from the TCGA database (https://portal.gdc.cancer.gov/). The overall survival rate graph of NSCLC is from the study conducted by Mao et al., and the usage of the graph was approved (PMID: 30094095).
References
Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–99.
Boland M, Nazor K, Loring J. Epigenetic regulation of pluripotency and differentiation. Circulation Res. 2014;115:311–24.
Dobersch, S, Rubio, K, Barreto, G. Pioneer factors and architectural proteins mediating embryonic expression signatures in cancer. Trends Mol Med. 2019. https://doi.org/10.1016/j.molmed.2019.01.008.
Singh AK, Mueller-Planitz F. Nucleosome positioning and spacing: from mechanism to function. J Mol Biol. 2021;433:166847.
Flaus A, Martin D, Barton G, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–905.
Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem. 2014;83:671–96.
Ozturk N, Singh I, Mehta A, Braun T, Barreto G. HMGA proteins as modulators of chromatin structure during transcriptional activation. Front Cell Dev Biol. 2014;2:5.
Lee DW, Zhang K, Ning ZQ, Raabe EH, Tintner S, Wieland R, et al. Proliferation-associated SNF2-like gene (PASG): a SNF2 family member altered in leukemia. Cancer Res. 2000;60:3612–22.
Jarvis CD, Geiman T, Vila-Storm MP, Osipovich O, Akella U, Candeias S, et al. A novel putative helicase produced in early murine lymphocytes. Gene. 1996;169:203–7.
Geiman T, Tessarollo L, Anver M, Kopp J, Ward J, Muegge K. Lsh, a SNF2 family member, is required for normal murine development. Biochimica et Biophysica Acta. 2001;1526:211–20.
Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 2001;15:2940–4.
Yan Q, Huang J, Fan T, Zhu H, Muegge K. Lsh, a modulator of CpG methylation, is crucial for normal histone methylation. EMBO J. 2003;22:5154–62.
Muegge K, Geiman T, Xi S, Jiang Q, Schmidtman A, Chen T, et al. Lsh is involved in de novo methylation of DNA. Proc Am Assoc Cancer Res Annu Meet. 2006;47:541–541.
Zocchi L, Mehta A, Wu SC, Wu J, Gu Y, Wang J, et al. Chromatin remodeling protein HELLS is critical for retinoblastoma tumor initiation and progression. Oncogenesis. 2020;9:25.
Sha K, Boyer LA. The chromatin signature of pluripotent cells (May 31, 2009), StemBook, ed. The Stem Cell Research Community, StemBook, https://doi.org/10.3824/stembook.1.45.1, http://www.stembook.org.
Delgado-Olguin P, Recillas-Targa F. Chromatin structure of pluripotent stem cells and induced pluripotent stem cells. Brief Funct Genomics. 2011;10:37–49.
Ren J, Briones V, Barbour S, Yu W, Han Y, Terashima M, et al. The ATP binding site of the chromatin remodeling homolog Lsh is required for nucleosome density and de novo DNA methylation at repeat sequences. Nucleic Acids Res. 2015;43:1444–55.
Law C-T, Wei L, Tsang FH-C, Chan CY-K, Xu IM-J, Lai RK-H, et al. HELLS regulates chromatin remodeling and epigenetic silencing of multiple tumor suppressor genes in human hepatocellular carcinoma. Hepatology. 2019;69:2013–30.
Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22.
Mehta A, Dobersch S, Romero-Olmedo AJ, Barreto G. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev. 2015. https://doi.org/10.1007/s10555-015-9563-3.
Smith Z, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–20.
Jenness C, Giunta S, Müller MM, Kimura H, Muir TW, Funabiki H. HELLS and CDCA7 comprise a bipartite nucleosome remodeling complex defective in ICF syndrome. Proc Natl Acad Sci USA. 2018;115:E876–e885.
Termanis A, Torrea N, Culley J, Kerr A, Ramsahoye B, Stancheva I. The SNF2 family ATPase LSH promotes cell-autonomous de novo DNA methylation in somatic cells. Nucleic Acids Res. 2016;44:7592–604.
De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat Cell Biol. 2006;8:1448–54.
Zeng W, Baumann C, Schmidtmann A, Honaramooz A, Tang L, Bondareva A, et al. Lymphoid-specific helicase (HELLS) is essential for meiotic progression in mouse spermatocytes. Biol Reprod. 2011;84:1235–41.
Fan T, Yan Q, Huang J, Austin S, Cho E, Ferris D, et al. Lsh-deficient murine embryonal fibroblasts show reduced proliferation with signs of abnormal mitosis. Cancer Res. 2003;63:4677–83.
Baumann C, Ma W, Wang X, Kandasamy MK, Viveiros MM, De La Fuente R. Helicase LSH/Hells regulates kinetochore function, histone H3/Thr3 phosphorylation and centromere transcription during oocyte meiosis. Nat Commun. 2020;11:4486–4486.
Dunican DS, Cruickshanks HA, Suzuki M, Semple CA, Davey T, Arceci RJ, et al. Lsh regulates LTR retrotransposon repression independently of Dnmt3b function. Genome Biol. 2013;14:R146.
Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–5.
Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 2017;18:517–34.
Ross SE, Bogdanovic O. TET enzymes, DNA demethylation and pluripotency. Biochem Soc Trans. 2019;47:875–85.
Jia J, Shi Y, Chen L, Lai W, Yan B, Jiang Y, et al. Decrease in lymphoid specific helicase and 5-hydroxymethylcytosine is associated with metastasis and genome instability. Theranostics. 2017;7:3920–32.
van der Wijst M, Venkiteswaran M, Chen H, Xu G, Plösch T, Rots M. Local chromatin microenvironment determines DNMT activity: from DNA methyltransferase to DNA demethylase or DNA dehydroxymethylase. Epigenetics. 2015;10:671–6.
Wang Y, Fischle W, Cheung W, Jacobs S, Khorasanizadeh S, Allis CD. Beyond the double helix: writing and reading the histone code. Novartis Found Symp. 2004;259:3–17. discussion 17–21, 163–169.
Singh I, Ozturk N, Cordero J, Mehta A, Hasan D, Cosentino C, et al. High mobility group protein-mediated transcription requires DNA damage marker gamma-H2AX. Cell Res. 2015;25:837–50.
Dobersch S, Rubio K, Singh I, Gunther S, Graumann J, Cordero J, et al. Positioning of nucleosomes containing gamma-H2AX precedes active DNA demethylation and transcription initiation. Nat Commun. 2021;12:1072.
Yu W, Briones V, Lister R, McIntosh C, Han Y, Lee EY, et al. CG hypomethylation in Lsh-/- mouse embryonic fibroblasts is associated with de novo H3K4me1 formation and altered cellular plasticity. Proc Natl Acad Sci USA. 2014;111:5890–5.
Ren J, Hathaway NA, Crabtree GR, Muegge K. Tethering of Lsh at the Oct4 locus promotes gene repression associated with epigenetic changes. Epigenetics. 2018;13:173–81.
Han Y, Ren J, Lee E, Xu X, Yu W, Muegge K. Lsh/HELLS regulates self-renewal/proliferation of neural stem/progenitor cells. Sci Rep. 2017;7:1136.
Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–56.
Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4.
Singh I, Contreras A, Cordero J, Rubio K, Dobersch S, Gunther S, et al. MiCEE is a ncRNA-protein complex that mediates epigenetic silencing and nucleolar organization. Nat Genet. 2018;50:990–1001.
Myant K, Stancheva I. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol. 2008;28:215–26.
Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017;168:644–56.
Unoki M, Funabiki H, Velasco G, Francastel C, Sasaki H. CDCA7 and HELLS mutations undermine nonhomologous end joining in centromeric instability syndrome. J Clin Investig. 2019;129:78–92.
Aguilera A, Garcia-Muse T. Causes of genome instability. Annu Rev Genet. 2013;47:1–32.
Scully R, Panday A, Elango R, Willis N. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 2019;20:698–714.
Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–96.
Dimitrova N, de Lange T. Cell cycle-dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of nonhomologous end joining (NHEJ) in G1 and resection-mediated inhibition of NHEJ in G2. Mol Cell Biol. 2009;29:5552–63.
Mahaney B, Meek K, Lees-Miller S. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochemical J. 2009;417:639–50.
Basenko E, Kamei M, Ji L, Schmitz R, Lewis Z. The LSH/DDM1 homolog MUS-30 is required for genome stability, but not for DNA methylation in Neurospora crassa. PLoS Genet. 2016;12:e1005790.
He YF, Ren JK, Xu XP, Ni K, Schwader A, Finney R, et al. Lsh/HELLS is required for B lymphocyte development and immunoglobulin class switch recombination. Proc Natl Acad Sci USA. 2020;117:20100–8.
Kollárovič G, Topping C, Shaw E, Chambers A. The human HELLS chromatin remodelling protein promotes end resection to facilitate homologous recombination and contributes to DSB repair within heterochromatin. Nucleic Acids Res. 2020;48:1872–85.
Syed A, Tainer J. The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu Rev Biochem. 2018;87:263–94.
Sartori A, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–14.
Paull T. Mechanisms of ATM activation. Annu Rev Biochem. 2015;84:711–38.
Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci USA. 2008;105:11200–5.
Li J, Stern DF. DNA damage regulates Chk2 association with chromatin. J Biol Chem. 2005;280:37948–56.
Kakarougkas A, Ismail A, Klement K, Goodarzi A, Conrad S, Freire R, et al. Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res. 2013;41:9719–31.
Burrage J, Termanis A, Geissner A, Myant K, Gordon K, Stancheva I. The SNF2 family ATPase LSH promotes phosphorylation of H2AX and efficient repair of DNA double-strand breaks in mammalian cells. J cell Sci. 2012;125:5524–34.
Spruce C, Dlamini S, Ananda G, Bronkema N, Tian H, Paigen K, et al. HELLS and PRDM9 form a pioneer complex to open chromatin at meiotic recombination hot spots. Genes Dev. 2020;34:398–412.
Roos W, Krumm A. The multifaceted influence of histone deacetylases on DNA damage signalling and DNA repair. Nucleic Acids Res. 2016;44:10017–30.
Miller K, Tjeertes J, Coates J, Legube G, Polo S, Britton S, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–51.
Johnson D, Spitz G, Tharkar S, Quayle S, Shearstone J, Jones S, et al. HDAC1,2 inhibition impairs EZH2- and BBAP-mediated DNA repair to overcome chemoresistance in EZH2 gain-of-function mutant diffuse large B-cell lymphoma. Oncotarget. 2015;6:4863–87.
Rubio K, Singh I, Dobersch S, Sarvari P, Gunther S, Cordero J, et al. Inactivation of nuclear histone deacetylases by EP300 disrupts the MiCEE complex in idiopathic pulmonary fibrosis. Nat Commun. 2019;10:2229.
Lee SJ, Jang H, Park C. Maspin increases Ku70 acetylation and Bax-mediated cell death in cancer cells. Int J Mol Med. 2012;29:225–30.
Yang R, Liu N, Chen L, Jiang Y, Shi Y, Mao C, et al. GIAT4RA functions as a tumor suppressor in non-small cell lung cancer by counteracting Uchl3-mediated deubiquitination of LSH. Oncogene. 2019;38:7133–45.
Yang R, Liu N, Chen L, Jiang Y, Shi Y, Mao C, et al. LSH interacts with and stabilizes GINS4 transcript that promotes tumourigenesis in non-small cell lung cancer. J Exp Clin Cancer Res: CR. 2019;38:280.
Waseem A, Ali M, Odell E, Fortune F, Teh M. Downstream targets of FOXM1: CEP55 and HELLS are cancer progression markers of head and neck squamous cell carcinoma. Oral Oncol. 2010;46:536–42.
Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang Y, et al. A G3BP1-interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res. 2018;78:3484–96.
Chen L, Shi Y, Liu N, Wang Z, Yang R, Yan B, et al. DNA methylation modifier LSH inhibits p53 ubiquitination and transactivates p53 to promote lipid metabolism. Epigenetics Chromatin 2019;12:1–22.
Peng X, Sun J, Long Y, Xiao D, Zhou J, Tao Y, et al. The significance of HOXC11 and LSH in survival prediction in gastric adenocarcinoma. OncoTargets Ther. 2021;14:1517–29.
Yang R, Liu G, Han L, Qiu Y, Wang L, Wang M. MiR-365a-3p-mediated regulation of HELLS/GLUT1 axis suppresses aerobic glycolysis and gastric cancer growth. Front Oncol. 2021;11:616390.
Long J, Xia A, Liu J, Jing J, Wang Y, Qi C, et al. Decrease in DNA methylation 1 (DDM1) is required for the formation of CHH islands in maize. J Integr Plant Biol. 2019;61:749–64.
Jiang Y, Mao C, Yang R, Yan B, Shi Y, Liu X, et al. EGLN1/c-Myc induced lymphoid-specific helicase inhibits ferroptosis through lipid metabolic gene expression changes. Theranostics. 2017;7:3293–305.
Liu Y, Mao C, Wang M, Liu N, Ouyang L, Liu S, et al. Cancer progression is mediated by proline catabolism in non-small cell lung cancer. Oncogene. 2020;39:2358–76.
Dixon S, Stockwell B. The role of iron and reactive oxygen species in cell death. Nat Chem Biol. 2014;10:9–17.
Liu, N, Lin, X, Huang, C. Activation of the reverse transsulfuration pathway through NRF2/CBS confers erastin-induced ferroptosis resistance. Br J Cancer. 2019. https://doi.org/10.1038/s41416-019-0660-x.
Myant K, Termanis A, Sundaram A, Boe T, Li C, Merusi C, et al. LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res. 2011;21:83–94.
He X, Yan B, Liu S, Jia J, Lai W, Xin X, et al. Chromatin remodeling factor LSH drives cancer progression by suppressing the activity of fumarate hydratase. Cancer Res. 2016;76:5743–55.
von Eyss B, Maaskola J, Memczak S, Möllmann K, Schuetz A, Loddenkemper C, et al. The SNF2-like helicase HELLS mediates E2F3-dependent transcription and cellular transformation. EMBO J. 2012;31:972–85.
Rawłuszko-Wieczorek A, Siera A, Jagodziński P. TET proteins in cancer: current ‘state of the art’. Crit Rev Oncol/Hematol. 2015;96:425–36.
Spruijt C, Gnerlich F, Smits A, Pfaffeneder T, Jansen P, Bauer C, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–59.
Liu N, Yang R, Shi Y, Chen L, Liu Y, Wang Z, et al. The cross-talk between methylation and phosphorylation in lymphoid-specific helicase drives cancer stem-like properties. Signal Transduct Target Ther. 2020;5:197.
Chen D, Maruschke M, Hakenberg O, Zimmermann W, Stief C, Buchner A. TOP2A, HELLS, ATAD2, and TET3 are novel prognostic markers in renal cell carcinoma. Urology. 2017;102:265.e261–265.e267.
Yan X, Dong X, Liu L, Yang Y, Lai J, Guo Y. DNA methylation signature of intergenic region involves in nucleosome remodeler DDM1-mediated repression of aberrant gene transcriptional read-through. J Genet genomics = Yi Chuan Xue Bao. 2016;43:513–23.
Corem S, Doron-Faigenboim A, Jouffroy O, Maumus F, Arazi T, Bouché N. ddm1Redistribution of CHH methylation and small interfering RNAs across the Genome of Tomato Mutants. Plant cell. 2018;30:1628–44.
Ito T, Tarutani Y, To T, Kassam M, Duvernois-Berthet E, Cortijo S, et al. Genome-wide negative feedback drives transgenerational DNA methylation dynamics in Arabidopsis. PLoS Genet. 2015;11:e1005154.
Kinoshita Y, Saze H, Kinoshita T, Miura A, Soppe W, Koornneef M, et al. Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J: Cell Mol Biol. 2007;49:38–45.
Zemach A, Kim M, Hsieh P, Coleman-Derr D, Eshed-Williams L, Thao K, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205.
Lyons, D, Zilberman, D. DDM1 and Lsh remodelers allow methylation of DNA wrapped in nucleosomes. eLife. 2017;6:e30674.
Tan F, Lu Y, Jiang W, Wu T, Zhang R, Zhao Y, et al. DDM1 represses noncoding RNA expression and RNA-directed DNA methylation in heterochromatin. Plant Physiol. 2018;177:1187–97.
Wang D, Qu Z, Yang L, Zhang Q, Liu Z, Do T, et al. Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants. Plant J: Cell Mol Biol. 2017;90:133–46.
Litwin I, Bakowski T, Maciaszczyk-Dziubinska E, Wysocki R. The LSH/HELLS homolog Irc5 contributes to cohesin association with chromatin in yeast. Nucleic Acids Res. 2017;45:6404–16.
Mao C, Wang M, Qian B, Ouyang L, Shi Y, Liu N, et al. Aryl hydrocarbon receptor activated by benzo (a) pyrene promotes SMARCA6 expression in NSCLC. Am J Cancer Res. 2018;8:1214–27.
Acknowledgements
We thank all the members of the laboratory for their resourceful comments on the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [82072594, 81672787, YT; 82073097, 81874139, 81672991, S.Liu; 82002916, CM; 82073136, 81772927, DX], China Postdoctoral Science Foundation [2019 M652804, CM], Natural Science Foundation of Hunan Province [2020JJ5790, CM], Hunan Provincial Key Area R&D Programs [2019SK2253, YT], Postdoctoral Foundation of Central South University [220372, CM], Shenzhen Science and Technology Program [KQTD20170810160226082, YT], and Shenzhen Municipal Government of China [JCYJ20180507184647104, YT]. Guillermo Barreto was funded by the “Université Paris-Est Créteil” (UPEC, Créteil, France), the “Centre National de la Recherche Scientifique” (CNRS, France), “Délégation Centre-Est” (CNRS-DR6), the “Lorraine Université” (LU, France) through the initiative “Lorraine Université d’Excellence” (LUE) and the dispositive “Future Leader” and the “Deutsche Forschungsgemeinschaft” (DFG, Bonn, Germany) (BA 4036/4-1). Karla Rubio was funded by the “Consejo de Ciencia y Tecnología del Estado de Puebla” (CONCYTEP, Puebla, Mexico) through the initiative International Laboratory EPIGEN.
Author information
Authors and Affiliations
Contributions
YT, DX and GB contributed to the conception of the study; XC, Yamei Li and KR wrote the manuscript; BD, Yuyi Li and QT performed the data analyses; CM and SL helped with constructive discussions.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This work does not require any ethical approval or participating consent.
Consent to publish
Not applicable.
Competing interests
The authors declare no competing interests. This manuscript has been read and approved by all the authors, and has not been submitted to or is not under consideration for publication elsewhere.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, X., Li, Y., Rubio, K. et al. Lymphoid-specific helicase in epigenetics, DNA repair and cancer. Br J Cancer 126, 165–173 (2022). https://doi.org/10.1038/s41416-021-01543-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01543-2
This article is cited by
-
The epitranscriptome of high-grade gliomas: a promising therapeutic target with implications from the tumor microenvironment to endogenous retroviruses
Journal of Translational Medicine (2023)
-
Unaltered hepatic wound healing response in male rats with ancestral liver injury
Nature Communications (2023)
-
USP11-mediated LSH deubiquitination inhibits ferroptosis in colorectal cancer through epigenetic activation of CYP24A1
Cell Death & Disease (2023)
-
GPR162 activates STING dependent DNA damage pathway as a novel tumor suppressor and radiation sensitizer
Signal Transduction and Targeted Therapy (2023)
-
PCDHB14 promotes ferroptosis and is a novel tumor suppressor in hepatocellular carcinoma
Oncogene (2022)