Abstract
Background
Individualising treatment in breast cancer requires effective predictive biomarkers. While relatively few genomic aberrations are clinically relevant, there is a need for characterising patients across different subtypes to identify actionable alterations.
Methods
We identified genomic alterations in 49 potentially actionable genes for which drugs are available either clinically or via clinical trials. We explored the landscape of mutations and copy number alterations (CNAs) in actionable genes in seven breast cancer subtypes utilising The Cancer Genome Atlas. To dissect the genomic complexity, we analysed the patterns of co-occurrence and mutual exclusivity in actionable genes.
Results
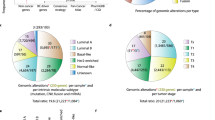
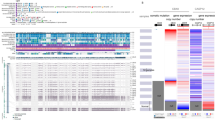
We found that >30% of tumours harboured putative actionable events that are targetable by currently available drugs. We identified genes that had multiple targetable alterations, representing candidate targets for combination therapy. Genes predicted to be drivers in primary breast tumours fell into five categories: mTOR pathway, immune checkpoints, oestrogen signalling, tumour suppression and DNA damage repair. Our analysis also revealed that CNAs in 34/49 (69%) and mutations in 13/49 (26%) genes were significantly associated with gene expression, validating copy number events as a dominant oncogenic mechanism in breast cancer.
Conclusion
These results may enable the acceleration of personalised therapy and improve clinical outcomes in breast cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data analysed in this study are available in the TCGA GDC data portal and cBioportal platform.
References
Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. https://doi.org/10.1038/nm.2323
Spitale A, Mazzola P, Soldini D, Mazzucchelli L, Bordoni A. Breast cancer classification according to immunohistochemical markers: clinicopathologic features and short-term survival analysis in a population-based study from the South of Switzerland. Ann Oncol. 2009;20:628–35. https://doi.org/10.1093/annonc/mdn675
Tang P, Wang J, Bourne P. Molecular classifications of breast carcinoma with similar terminology and different definitions: are they the same? Hum Pathol. 2008;39:506–13. https://doi.org/10.1016/j.humpath.2007.09.005
Li CI, Anderson BO, Daling JR, Moe RE. Trends in incidence rates of invasive lobular and ductal breast carcinoma. J Am Med Assoc. 2003;289:1421–4.
Tyanova S, Albrechtsen R, Kronqvist P, Cox J, Mann M, Geiger T. Proteomic maps of breast cancer subtypes. Nat Commun. 2016;7:10259 https://doi.org/10.1038/ncomms10259
Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. https://doi.org/10.1038/35021093
Harris LN, Ismaila N, McShane LM, Andre F, Collyar DE, Gonzalez-Angulo AM, et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2016;34:1134–50. https://doi.org/10.1200/jco.2015.65.2289
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. https://doi.org/10.1038/nature12477
Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971 https://doi.org/10.1038/ncomms9971
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. https://doi.org/10.1038/nature10983
Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–33. https://doi.org/10.1038/ng.2762
Cardoso F, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rubio IT, et al. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann Oncol. 2019;30:1194–220. https://doi.org/10.1093/annonc/mdz173
Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18–43. https://doi.org/10.1043/1543-2165(2007)131[18:Asocco]2.0.Co;2
Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–19. https://doi.org/10.1016/j.cell.2015.09.033
Kaur P, Porras TB, Ring A, Carpten JD, Lang JE. Comparison of TCGA and GENIE genomic datasets for the detection of clinically actionable alterations in breast cancer. Sci Rep. 2019;9:1482 https://doi.org/10.1038/s41598-018-37574-8
Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–8. https://doi.org/10.1038/nm.3559
Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–63. https://doi.org/10.1038/nbt.3391
Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–56. https://doi.org/10.1101/gr.239244.118
Mularoni L, Sabarinathan R, Deu-Pons J, Gonzalez-Perez A, López-Bigas N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 2016;17:128 https://doi.org/10.1186/s13059-016-0994-0
Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precision Oncol. 2017. https://doi.org/10.1200/po.17.00011.
Franch-Expósito S, Bassaganyas L, Vila-Casadesús M, Hernández-Illán E, Esteban-Fabró R, Díaz-Gay M, et al. CNApp, a tool for the quantification of copy number alterations and integrative analysis revealing clinical implications. eLife 2020;9. https://doi.org/10.7554/eLife.50267.
Deng M, Bragelmann J, Schultze JL, Perner S. Web-TCGA: an online platform for integrated analysis of molecular cancer data sets. BMC Bioinforma. 2016;17:72 https://doi.org/10.1186/s12859-016-0917-9
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. https://doi.org/10.1038/gim.2015.30
Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. https://doi.org/10.1016/j.cell.2012.04.024
Mayakonda A, Koeffler HP. Maftools: efficient analysis, visualization and summarization of MAF files from large-scale cohort based cancer studies. bioRxiv:052662,2016 [Preprint]. 2016. Available from: https://doi.org/10.1101/052662.
Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics. 2013;29:2238–44. https://doi.org/10.1093/bioinformatics/btt395
Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34:427–.e426. https://doi.org/10.1016/j.ccell.2018.08.008
AACR Project GENIE: powering precision medicine through an International Consortium. Cancer Discov. 2017; 7:818, https://doi.org/10.1158/2159-8290.CD-17-0151.
Angus L, Smid M, Wilting SM, van Riet J, Van Hoeck A, Nguyen L, et al. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat Genet. 2019;51:1450–8. https://doi.org/10.1038/s41588-019-0507-7
Lefebvre C, Bachelot T, Filleron T, Pedrero M, Campone M, Soria J-C, et al. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med. 2016;13:e1002201 https://doi.org/10.1371/journal.pmed.1002201
Spoerke JM, Gendreau S, Walter K, Qiu J, Wilson TR, Savage H, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016;7:11579 https://doi.org/10.1038/ncomms11579
Bergamaschi A, Kim YH, Wang P, Sørlie T, Hernandez-Boussard T, Lonning PE, et al. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes, Chromosomes Cancer. 2006;45:1033–40. https://doi.org/10.1002/gcc.20366
Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. https://doi.org/10.1038/nature11412
Weigman VJ, Chao HH, Shabalin AA, He X, Parker JS, Nordgard SH, et al. Basal-likebreast cancer DNA copy number losses identify genes involved in genomic instability, response to therapy, and patient survival. Breast Cancer Res Treat. 2012;133:865–80. https://doi.org/10.1007/s10549-011-1846-y
Tsimberidou AM, Kurzrock R. Precision medicine: lessons learned from the SHIVA trial. Lancet Oncol. 2015;16:e579–580. https://doi.org/10.1016/s1470-2045(15)00397-6
Boca SM, Kinzler KW, Velculescu VE, Vogelstein B, Parmigiani G. Patient-oriented gene set analysis for cancer mutation data. Genome Biol. 2010;11:R112 https://doi.org/10.1186/gb-2010-11-11-r112
Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012;22:398–406. https://doi.org/10.1101/gr.125567.111
Kim JW, Botvinnik OB, Abudayyeh O, Birger C, Rosenbluh J, Shrestha Y, et al. Characterizing genomic alterations in cancer by complementary functional associations. Nat Biotechnol. 2016;34:539–46. https://doi.org/10.1038/nbt.3527
Park S, Lehner B. Cancer type-dependent genetic interactions between cancer driver alterations indicate plasticity of epistasis across cell types. Mol Syst Biol. 2015;11:824–824. https://doi.org/10.15252/msb.20156102
Vandin F, Upfal E, Raphael BJ. De novo discovery of mutated driver pathways in cancer. Genome Res. 2012;22:375–85. https://doi.org/10.1101/gr.120477.111
Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19:495–509. https://doi.org/10.1038/s41568-019-0179-8
Muller FL, Colla S, Aquilanti E, Manzo VE, Genovese G, Lee J, et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488:337–42. https://doi.org/10.1038/nature11331
O’Shaughnessy J, Osborne C, Pippen J, Yoffe M, Patt D, Monaghan G, et al. Efficacy of BSI-201, a poly (ADP-ribose) polymerase-1 (PARP1) inhibitor, in combination with gemcitabine/carboplatin (G/C) in patients with metastatic triple-negative breast cancer (TNBC): results of a randomized phase II trial. J Clin Oncol. 2009;27:3–3. https://doi.org/10.1200/jco.2009.27.18_suppl.3
Tutt A, Robson M, Garber JE, Domchek S, Audeh MW, Weitzel JN, et al. Phase II trial of the oral PARP inhibitor olaparib in BRCA-deficient advanced breast cancer. J Clin Oncol. 2009;27:CRA501–CRA501. https://doi.org/10.1200/jco.2009.27.18_suppl.cra501
Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. https://doi.org/10.1056/NEJMoa0900212
Cisowski J, Sayin VI, Liu M, Karlsson C, Bergo MO. Oncogene-induced senescence underlies the mutual exclusive nature of oncogenic KRAS and BRAF. Oncogene. 2016;35:1328–33. https://doi.org/10.1038/onc.2015.186
Etemadmoghadam D, Weir BA, Au-Yeung G, Alsop K, Mitchell G, George J, et al. Synthetic lethality between CCNE1 amplification and loss of BRCA1. Proc Natl Acad Sci USA. 2013;110:19489–94. https://doi.org/10.1073/pnas.1314302110
Loibl S, von Minckwitz G, Schneeweiss A, Paepke S, Lehmann A, Rezai M, et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (her2) therapy in primary HER2-overexpressing breast cancer. J Clin Oncol. 2014;32:3212–20. https://doi.org/10.1200/jco.2014.55.7876
Levinson AD. Cancer therapy reform. Science. 2010;328:137 https://doi.org/10.1126/science.1189749
Fujimoto Y, Morita TY, Ohashi A, Haeno H, Hakozaki Y, Fujii M, et al. Combination treatment with a PI3K/Akt/mTOR pathway inhibitor overcomes resistance to anti-HER2 therapy in PIK3CA-mutant HER2-positive breast cancer cells. Sci Rep. 2020;10:21762 https://doi.org/10.1038/s41598-020-78646-y
André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380:1929–40. https://doi.org/10.1056/NEJMoa1813904
Tan DS, Thomas GV, Garrett MD, Banerji U, de Bono JS, Kaye SB, et al. Biomarker-driven early clinical trials in oncology: a paradigm shift in drug development. Cancer J. 2009;15:406–20. https://doi.org/10.1097/PPO.0b013e3181bd0445
Chow L, Sun Y, Jassem J, Baselga, J, Hayes D, Wolff A, et al. Phase 3 study of temsirolimus with letrozole or letrozole alone in postmenopausal women with locally advanced or metastatic breast cancer. Proc Breast Cancer Res Treat. 2006;S286.
Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–46. https://doi.org/10.1038/nrd4504
Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 2010;29:4018–32. https://doi.org/10.1038/onc.2010.154
Acknowledgements
The authors wish to acknowledge the TCGA Research Network for sharing the TCGA breast cancer genomic datasets. The results presented here are in whole or part based upon data generated by TCGA managed by the NCI and NHGRI. We thank the USC Libraries Bioinformatics Service for assisting with data analysis. The bioinformatics software and computing resources used in the analysis are funded by the USC Office of Research and the Norris Medical Library.
Funding
The project was supported in part by award number P30CA014089 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institute of Health. The project was also supported by the Woodbury Foundation.
Author information
Authors and Affiliations
Contributions
Study design: PK and JEL; data acquisition: PK and JEL; data analysis: PK, JEL, AC, TP and AR; manuscript drafting: PK and JEL; critical revisions: PK, JEL, AR, TP, AC, JL and IK; funding: JEL.
Corresponding author
Ethics declarations
Competing interests
Dr. Kang is a member of Puma Biotechnology speaker bureau. The remaining authors declare no competing interests.
Ethics approval and consent to participate
The study was performed in strict accordance with the recommendations of data access guidelines of TCGA datasets. We received administrative permission for downloading the restricted-access data for breast cancer patients from the TCGA Data Access Committee (Project # 28198). The study was performed in accordance with the Declaration of Helsinki.
Consent to publish
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kaur, P., Porras, T.B., Colombo, A. et al. Identification of putative actionable alterations in clinically relevant genes in breast cancer. Br J Cancer 125, 1270–1284 (2021). https://doi.org/10.1038/s41416-021-01522-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01522-7