Abstract
Advances in genomic strategies and the development of targeted therapies have enabled precision medicine to revolutionise the field of oncology. Precision medicine uses patient-specific genetic and molecular information, traditionally obtained from tumour biopsy samples, to classify tumours and treat them accordingly. However, biopsy samples often fail to provide complete tumour profiling, and the technique is expensive and, of course, relatively invasive. Advances in genomic techniques have led to improvements in the isolation and detection of circulating tumour DNA (ctDNA), a component of a peripheral blood draw/liquid biopsy. Liquid biopsy offers a minimally invasive method to gather genetic information that is representative of a global snapshot of both primary and metastatic sites and can thereby provide invaluable information for potential targeted therapies and methods for tumour surveillance. However, a lack of prospective clinical trials showing direct patient benefit has limited the implementation of liquid biopsies in standard clinical applications. Here, we review the potential of ctDNA obtained by liquid biopsy to revolutionise personalised medicine and discuss current applications of ctDNA both at the benchtop and bedside.
Similar content being viewed by others
Background
Advances in methods to identify and detect tumour-specific molecular biomarkers over the past two decades have led to an increased potential for personalised treatment. The focus on precision medicine in the field of oncology has vastly improved the development of targeted therapies and overall patient outcome1,2. Moreover, despite the initial success of targeted therapies, cancers often develop distinct mechanisms of resistance and patients eventually relapse, making the need to detect and monitor molecular biomarkers in response to treatment important. Although tissue/tumour biopsy samples remain the gold standard from which molecular information is gathered, the presence of tumour heterogeneity or multiple tumour sites presents a significant challenge. Furthermore, tissue biopsies can be expensive, invasive, and difficult to perform3. The development of liquid biopsies, and advances in the detection of circulating tumour DNA (ctDNA)—fragments of DNA derived from tumour cells—have provided clinicians with an effective method to address the shortcomings associated with traditional biopsy samples.
Liquid biopsy samples refer to harvested bodily fluids, often blood, and the associated components (RNA, DNA, circulating tumour cells, extracellular vesicles, etc.), which provide information on potential biomarkers4. In this review, we will focus specifically on the potential clinical use of ctDNA detected in liquid biopsy samples. The presence of ctDNA in liquid biopsy samples enables clinicians to serially collect and quantify genetic information from multiple tumour sites. Consequently, this benefit, among others, has led to an explosion in research to determine the many potential applications of liquid biopsy in screening, prognostics, treatment monitoring, and clinical decision making, and a number of FDA-approved techniques that assess ctDNA are currently in use (Box 1).
However, despite the numerous potential benefits of ctDNA-based liquid biopsy, several ongoing concerns exist. For example, the mutational profiles obtained from tumour biopsy samples are not always concordant with those present in ctDNA, which potentially raises concerns about sensitivity and specificity5,6. These discrepancies often result from the low levels of tumour-derived mutations in blood, along with the presence of naturally occurring variants that are not associated with the tumour, including clonal haematopoiesis, in which blood cells derived from a single clone are overrepresented. The low abundance of ctDNA in the blood might also fail to provide an accurate representation of tumour heterogeneity. Sample processing issues can further exacerbate issues of low ctDNA concentration and contamination by non-cancer associated variants7,8,9. Unfortunately, a consensus on appropriate pre-analytical steps to alleviate these difficulties has yet to be determined. To address these overarching concerns, scientists are working to improve techniques used throughout ctDNA pipelines, including isolation, detection and analysis. However, until these issues are sufficiently addressed and benefit is demonstrated in clinical trials, the use of ctDNA-based tests is best limited to specific situations. Currently, qualitative mutation detection for personalised treatment choices, particularly for late-stage disease, is the most prominent clinical use. However, translational research laboratories and companies have demonstrated the increasing potential of ctDNA analysis in applications such as disease surveillance and identification, which could significantly impact the mortality rates of individual cancers.
Presently, the clinical utility of ctDNA-based liquid biopsy approaches is being heavily explored in clinical breast cancer research. Breast cancer genetics, genomics and mutational profiles have been well-studied and defined over the years, and researchers are focusing on using ctDNA in conjunction with additional diagnostic tests in an effort to improve detection and treatment. Furthermore, the multiple subtypes of breast cancer each have their own molecular targets with well-known resistance mutations, for which ctDNA analysis could inform treatment decisions. In this review we seek to highlight commonly used ctDNA detection methods, survey the literature on the use of ctDNA in breast cancer and delineate areas of continuing development in ctDNA-based liquid biopsy towards providing personalised care for patients with breast cancer.
Sample processing
The extraction and isolation of cell free DNA from plasma is an important step in the ability to utilise liquid biopsies for genetic information. Improper sample processing can lead to potential contamination from the rapid lysis of white blood cells. Although EDTA collection tubes may be used, they require sample processing within 6 h of sample collection or the sample is compromised. Streck Cell-Free DNA BCT collection tubes allow for stabile storage blood for up to 14 days at room temperature and are ideal for circulating free DNA (cfDNA) isolation10. Blood is centrifuged to separate the plasma, which contains ctDNA, and the buffy coat, which contains germline DNA11. Plasma is centrifuged a second time to remove cellular contaminants and processed using a circulating nucleic acid isolation kit (i.e. QIAamp® Circulating Nucleic Acid Kit). The sample is then ready to be analysed. An overview of sample processing for ctDNA is shown in Fig. 1.
Blood samples are collected in tubes made specifically to stabilize all cfDNA. The collected sample is then centrifuged and cfDNA-containing plasma is isolated. Remaining cellular contamination is eliminated with a final centrifugation, and cfDNA is isolated and purified through a variety of commercial kits.
Measuring ctDNA obtained by liquid biopsy
Techniques for measuring cfDNA were first introduced in the early 1990s with a technique that utilised allele-specific PCR12. As technologies for detecting cfDNA expanded, they developed into two distinct but broad categories: PCR-based techniques and next generation sequencing (NGS)-based techniques. These techniques each have their benefits and disadvantages. Below is a broad overview of a few cfDNA technologies to date (Fig. 2). It is important to note that the list of ctDNA technologies is expansive and new technologies are constantly being developed.
Cell-free DNA is primarily analyzed for ctDNA content by PCR-based techniques or NGS-based techniques. In the PCR-based techniques few variants are probed at once whereas in NGS-based techniques many genes are queried in one sequencing run. Each technique has benefits and drawbacks regarding time, money, breadth, sensitivity, and scalability. Images were adapted from literature cited in the text.
PCR-based techniques
PCR-based techniques—including real-time quantitative PCR (qPCR), Beads, Emulsion, Amplifying and Magnetics (BEAMing) and droplet-digital PCR (ddPCR)—offer the simplest approach for the detection and quantification of ctDNA. PCR-based methods generally use target-specific probes that are designed to identify single-nucleotide variants. Thus, only known variants can be queried, and only a few variants can be probed at one time. Consequently, these techniques are typically applied to detect mutations that are highly prevalent or to track previously verified mutations.
qPCR
qPCR is a rapid and cheap method that most often uses a quenched fluorescent probe, which is released during amplification and emits a detectable fluorescent signal. Although the specificity of qPCR is quite high, significant variation can occur between repeated runs13, which has led to the development of new qPCR-based techniques to improve assay performance. However, these detection techniques have a relatively low threshold of mutation detection (0.1%) and cannot accurately detect extremely rare variants14,15.
BEAMing
BEAMing is a digital PCR method that combines the techniques of PCR and flow cytometry16. Magnetic beads are tagged with bait DNA and mixed with oil, PCR reagents and target DNA to form an emulsion. Each oil droplet is capable of undergoing PCR-mediated amplification of the DNA that is bound to the bait DNA. The emulsions are then magnetically purified and opened to isolate the amplified target DNA, which is identified with hybridised fluorescent probes and enumerated by flow cytometry. BEAMing has been reported to detect 1 mutant DNA molecule among 10,000 normal molecules, which is 10× more sensitive than improved qPCR assays17. However, despite the improved sensitivity, BEAMing is laborious and expensive, and only a few laboratories and commercial entities are capable of efficiently employing the technology.
ddPCR
The widely used technique of ddPCR incorporates the methodology of both qPCR and BEAMing18. ddPCR involves an emulsion of quenched fluorescent DNA probes, PCR components and sample DNA. Theoretically, the emulsion is diluted to contain ≤1 DNA molecule per oil droplet. PCR amplification removes the probe’s quencher, and fluorescent droplets are quantified using flow cytometry. Compared to BEAMing and qPCR, ddPCR is a relatively inexpensive method and offers improved performance metrics13,19,20.
Next generation sequencing (NGS)-based techniques
Next-generation sequencing (NGS) is technically complicated but offers the ability to multiplex samples in a single run, thereby significantly improving scalability and allowing for the detection of rare or unknown mutations—that is, a priori knowledge of the mutation is not required. The inner workings of NGS are beyond the scope of this review and have been discussed elsewhere21,22. Generally, however, NGS in the context of ctDNA evaluation involves sequencing the exons and the intron/exon junctions of a subset of cancer-related genes to identify both known and unknown mutations. Although this approach can detect mutant alleles at frequencies as low as 0.1%, many sequencing platforms have a random error rate of the same frequency23. Therefore, standard versions of this technique might not be sufficient for analysing low amounts of ctDNA, such as those in early cancer patients or post-surgery. However, advanced barcoding systems, improved hybrid capture techniques, and/or greater sequencing read depth have been implemented to minimise errors and maximise detection.
The first such technology to be developed was dubbed ‘Safe-Seqs’ for Safe-Sequencing System24. Additional iterations, including duplex sequencing25 and Targeted Error Correction Sequencing (TEC-seq)26, among others, ensued. All of these techniques use barcoded cfDNA sequencing libraries to generate redundant sequences of the same barcoded molecule. For clarity, cfDNA refers to all the DNA that is derived from a liquid biopsy—that is, DNA that originates from both normal and cancer cells—whereas ctDNA refers to DNA originating from cancer cells only (thus, ctDNA is a subset of cfDNA). These cfDNA molecules can then be consolidated to determine if any mutations found are bona fide mutations or whether they are due to sequencing errors, as true mutations should be present in multiple NGS reads containing the same barcodes. Integrated Digital Error Suppression for CAncer Personalised Profiling by deep sequencing (iDES-CAPP-seq) involves both single-stranded and double-stranded molecular barcoding with in silico error suppression, which enables the results to be optimised for molecule recovery and to detect sequencing mistakes, respectively27.
The development of these technologies significantly improves the sensitivity of NGS-based techniques while enabling the interrogation of thousands of genomic positions at once. Although promising, the clinical validity of NGS-based techniques for the detection of ctDNA still requires further evaluation, and their clinical utility—that is, whether these tests can change outcomes to help guide care for breast cancer patients—has yet to be demonstrated.
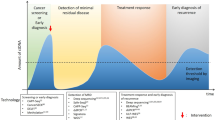
ctDNA IN EARLY DISEASE DETECTION
The early detection of breast cancer is critical to patient outcome: detection at the localised stage confers a 99% 5-year relative survival rate compared with a 27% 5-year relative survival rate for distant metastases28. Currently, mammography is the primary screening mode for breast cancer and is recommended by all leading medical organisations for women of average risk between the ages of 50 and 75 years. However, overdiagnosis and false positives can be associated with mammography29. Similar issues have been reported for screening modalities for other cancer types, such as prostate-specific antigen (PSA) screens for prostate cancer. On the basis of the potential survival benefits derived from early detection, research groups have therefore sought to design more accurate methods of detection for early-stage disease. Currently, many approaches to detect ctDNA are being studied to determine their role(s) in early disease detection (Table 1).
Total cfDNA levels
One of the simplest approaches used to assess early stages of disease is the analysis of total cfDNA levels. Cancer patients have higher levels of total cfDNA (comprising DNA form normal and cancer cells) compared with control populations, typically determined using qPCR or a fluorescent DNA quantification assay. Agassi et al. used a fluorescent assay to confirm that patients with early-stage breast cancer had levels of cfDNA that were higher, pre-surgery, than those measured in patients post-surgery, in healthy patients or in patients with pre-cancerous lesions. This method outperformed the blood-based breast cancer protein biomarker CA15-3, as the sensitivity and specificity of increased levels of cfDNA to detect breast cancer were 72% and 75% respectively while CA15-3 only identified 11% of patients. Notably, sensitivity of the cfDNA assay correlated with later stage disease, identifying 100% of patients with lymph node involvement30. Another study using cfDNA as a biomarker demonstrated that a qPCR-based assay could correctly identify 89 out of 100 patients with early-stage breast cancer, with a false positive rate of only 6%31. To address the discrepancies in total cfDNA-based classifications, Yu et al. performed a meta-analysis, and determined that the pooled sensitivity and specificity of multiple techniques was 87%32. It is important to note, however, that elevated cfDNA levels are not exclusive to tumours and might be affected by other comorbidities, resulting in false positive results. The measurement of cfDNA levels has been explored as a possible adjunct test with mammography to decrease false positives, by distinguishing benign from malignant lesions, and thereby prevent overtreatment. However, Peled et al. reported that the assessment of total cfDNA in suspected breast cancer patients was unable to discriminate between benign and malignant lesions and that additional biomarkers might be required33.
ctDNA detection
Techniques to detect ctDNA specific mutations from cfDNA, including targeted PCR-based approaches and NGS, have vastly improved the use of cfDNA in cancer detection. Targeted techniques used for screening, such as BEAMing and ddPCR, are best applied to highly prevalent mutations in breast cancer. For example, Beaver et al. used ddPCR to detect hotspot PIK3CA mutations from within the blood of 14 out of 15 early-stage breast cancer patients with a confirmed PIK3CA mutation found in tumour tissues34. Although PIK3CA mutations occur only in approximately 45% of oestrogen-receptor-positive (ER+) breast cancers, the study demonstrates that ddPCR analysis is an effective targeted technique for detecting early-stage disease specifically within this population. However, digital PCR platforms can only query for known mutations, so more and more studies have begun to use NGS platforms for ctDNA detection in breast and other cancers. NGS approaches aim to increase the gene panel size and, therefore, increase mutation detection sensitivity. In a pan-cancer study, Bettegowda et al. were able to detect ctDNA in 50% of localised breast cancer cases through a combination of BEAMing and Safe-SeqS35. Currently, NGS panels used for ctDNA detection test a relatively small number of genes/genomic locations and might not include less common genetic variants that contribute to tumour phenotypes. Therefore, there remains a significant need for ctDNA detection technologies that maintain a high sequencing depth across a broad set of cancer genes, with acceptable turnaround times and costs.
ctDNA in presymptomatic detection of cancer
The use of ctDNA in detecting cancer at its earliest, pre-symptomatic, screening stage is also being studied. However, data analysing viral DNA in nasopharyngeal carcinomas suggest that ctDNA detection will require the development of additional technology to achieve the level of sensitivity and specificity necessary for a primary cancer screening test. In a prospectively enrolled clinical trial, qPCR was used to detect circulating Epstein Barr virus (EBV) DNA in asymptomatic patients before the onset of nasopharyngeal carcinoma. Certain nasopharyngeal carcinomas exhibit high levels of EBV DNA and, through gene amplification, DNA detection rates were high in these patients, with 97.1% sensitivity and 98.6% specificity. The authors of this study utilised 50 genomes of EBV, each with approximately 10 PCR target sites for their screen, suggesting that approximately 500 mutations/variants would be required to achieve similar sensitivity and specificity to more traditional mutation-based assays36. This number is currently higher than the number of genes surveyed by most ctDNA NGS and targeted techniques. Although not a study of breast cancer, these data highlight the need for additional cancer biomarkers to achieve ideal screening accuracy.
To overcome this barrier, researchers have explored the inclusion of other analytes, such as protein biomarkers, into ctDNA screening. CancerSEEK (Thrive Earlier Detection) is a technique that uses a panel of cfDNA amplicons and protein biomarkers37. For the detection of early-stage (stages I–III) ovarian and liver cancer, this technique achieved 99% sensitivity in a retrospective analysis; however, a 33% median sensitivity was observed for all breast cancers. It is important to note that the difference in sensitivity may be directly linked to stage. 76% of ovarian cancer cases studied were in stage III compared to only 30% in breast cancer. This may equate for the observed differences in sensitivity while still maintaining a high specificity for both. In the prospective DETECT-A (a pan-cancer detection study performed in approximately 10,000 women), CancerSEEK in combination with PET-CT was able to increase sensitivity and specificity compared to CancerSEEK alone. Importantly, the use of PET-CT allowed the investigators to identify/locate the tissue of origin from which the cancers arose. However, the results were highly variable depending on the cancer type, and the majority of breast cancers were initially detected by standard of care screening38. To expand these studies to other cancer types, with longer term follow-up, another prospective trial is currently being run (NCT04213326).
Methylation profiling
Methylated ctDNA profiling has gained attention as an alternative detection method for cancer and has the potential advantage of being able to identify the cancer type. Both targeted and untargeted studies have found that methylated gene promoters represent hundreds to thousands of changes in transformed cells and can serve as biomarkers for specific cancers, including breast cancer39,40. Shen et al. performed whole genome methylation analysis in a small cohort of early-stage breast cancer patients using the technique of cell-free Methylated DNA ImmunoPrecipitation sequencing (cfMeDIP-seq) to achieve an ~85% detection rate41. Currently, large clinical studies led by GRAIL Inc. (Illumina) are ongoing to determine the potential of whole genome methylation profiling for early detection (NCT03934866; NCT03085888; NCT04241796; NCT03372902). Preliminary reports indicate that 60% of early-stage hormone-receptor negative (HR–) and 18% of early-stage HR+ breast cancers can be detected with 99% specificity42. The results of further prospective trials will determine the clinical validity and utility of these pan-cancer screening methods to detect early-stage cancers.
ctDNA PROGNOSIS: EARLY INDICATIONS OF OUTCOME AND RECURRENCE
As well as investigating the use of liquid biopsy approaches for the detection of early disease, researchers have made strides towards predicting patient outcome through ctDNA testing. This approach could allow those patients who are at high risk to receive additional treatment to prevent recurrence, as well as limiting overtreatment for those at low risk. Other studies aim to detect recurrence as early as possible to implement aggressive therapy (Table 1). If successful, these strategies could prevent suffering associated with unnecessary treatment, as well as the onset of advanced disease.
Prognostication for early disease
The early measurement of ctDNA levels across breast cancer subtypes has demonstrated prognostic ability. This prognostic potential has been particularly well studied in treatment-naïve disease. Rothé et al. found that, in HER2+ breast cancers, a lack of detectable PIK3CA and TP53 variants in patient plasma prior to neoadjuvant therapy was associated with a high pCR (pathological complete response) rate43. Similarly, a separate study demonstrated that the prevalence of mutant PIK3CA ctDNA pre-surgery was associated with poor relapse-free survival and overall survival, regardless of subtype44.
The prognosis of early-stage disease using ctDNA is not limited to the detection of mutational variants in ctDNA. A small study of breast cancer patients demonstrated that detection in ctDNA of the 1q21.3 amplification pre-surgery was prognostic for relapse, regardless of subtype45. Additionally, studies involving ctDNA epigenetics determined that the presence of one specific methylated region pre-chemotherapy was a poor prognostic factor, and >70% of patients with this marker relapsed within 5 years46. Although these studies focused on prognostication in the pre-treatment setting, other investigators have studied ctDNA during neoadjuvant therapy. Studies in the neoadjuvant setting have shown that the levels of certain ctDNA variants measured quantitatively by ddPCR correlated directly (positively or negatively, depending on the variant) with disease-free survival and overall survival in patients with triple negative breast cancer47,48. McDonald et al. corroborated this result using a specialised sequencing technique49. While these studies indicate the power of ctDNA in prognosticating patient outcomes, no prospective clinical trial to date has validated the prognostic potential of ctDNA in early-stage disease. However, the identification and confirmation of high-risk groups through such analyses could lead to more intense treatment in a targeted high-risk population as well as de-escalation of therapy in low risk groups.
Detecting minimal residual disease and predicting subsequent relapse
The rate of breast cancer recurrence within 10 years of curative-intent therapy has been shown to be 36.8%50. The ability to accurately identify those patients in whom cancer will recur could lead to increased survival through earlier treatment and prevention. Preliminary studies by Beaver et al.34 demonstrated that ddPCR could detect mutant PIK3CA ctDNA in some early-stage breast cancer patients after surgery as well as before. In the 29 patient cohort, PIK3CA mutations were present in both tumour tissue and pre-surgery blood samples from 14 patients. Post-surgery, 5 of these 14 patients remained mutation-positive, as assessed by ddPCR, suggesting the presence of minimal residual disease (MRD), and raising the possibility that ctDNA abundance post-surgery could identify patients at risk of recurrence. Indeed, one patient with detectable ctDNA post-surgery had triple negative metaplastic breast cancer, a very aggressive form of the disease, and died within two years of her initial diagnosis. Garcia-Murillas et al.51 used ddPCR to track patient-specific ctDNA mutations in the plasma of patients with early-stage breast cancer. In this study, serial post-surgery ctDNA liquid biopsy identified 12 of 15 relapses and correctly classified 96% of non-relapsing patients. Similarly, Olsson et al. used ddPCR to demonstrate that levels of cancer-specific genetic rearrangements were associated with relapse52. Patient panels of 4–6 rearrangements were used to detect ctDNA retrospectively in 93% of patients in whom cancer recurred. Despite advances in ddPCR, however, this approach is limited, as mentioned above, by the number of genetic alterations that can be queried in a biased manner. In this regard, NGS overcomes this limitation owing to its ability to query multiple loci in an unbiased way. Accordingly, studies have leveraged NGS-based approaches I conjunction with liquid biopsies for the detection of ctDNA as a surrogate for MRD. Parsons et al. demonstrated that an NGS-based approach was capable of measuring 488 mutations with 100 times more power than a single mutation ddPCR test in vitro53. MRD was detected using a patient-specific version of this assay, and all patients in whom ctDNA was detected 1-year post treatment relapsed (6 of 6). However, many patients still relapsed without the detection of ctDNA at this timepoint, potentially owing to the occurrence of longer disease-free intervals in these patients. In a similar approach by Coombes et al.54, 49 breast cancer patients were monitored using ultradeep sequencing of 16 variants to detect ctDNA as an MRD biomarker associated with recurrence. ctDNA was detected in 16 of the 18 patients in whom recurrence occurred, as early as 2 years prior to relapse. These studies indicate that the frequency and duration of monitoring will be critical issues for prospective trials that are monitoring ctDNA as a prognostic marker for MRD and relapse. Additionally, these trials must also address how the detection of ctDNA might influence treatment escalation, which is currently being explored in c-TRAK TN (NCT03145961).
Opportunities for the treatment of metastatic disease
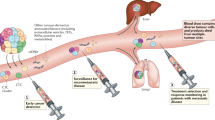
The levels of ctDNA have been demonstrated to directly reflect the tumour burden and stage of a patient’s cancer26, indicating that ctDNA also has the potential to facilitate real-time tumour surveillance in patients with active metastatic disease. This observation has led researchers to explore the possible clinical usefulness of ctDNA in monitoring the response to therapies in the metastatic setting (Table 1).
Monitoring and prognosticating late-stage disease progression
Serial plasma monitoring in patients with metastatic breast cancer gives clinicians the opportunity to quickly determine the response (if any) to current treatment. Multiple studies in metastatic breast cancer have demonstrated that a direct correlation exists between the levels of tumour-specific ctDNA and changes in diagnostic imaging seen in response to treatment. Although most of these studies primarily track the frequency of commonly mutated genes such as PIK3CA and TP5355,56,57, other alterations have also been tracked and yielded similar results45. Furthermore, these studies have demonstrated a link between the persistence of mutations in the ctDNA during ongoing therapy and poor outcome. According to additional studies, ctDNA dynamics during the early stages of treatment might also predict the clonal composition of the tumour upon progression. In the PALOMA-3 Study, a Phase 3 study of combination CDK4/6 inhibitor and fulvestrant, results suggest that early ctDNA dynamics demonstrate divergent response of tumour sub clones to treatment58. Recently, our group demonstrated that using cfDNA, we could accurately track non-random sub clonal dynamics in heterogenous tumours that significantly impacted the survival of cells59. The ability to identify these variants early-on could significantly impact the treatment course and potential response upon relapse. It is important to note, however, that such techniques are not yet useful for clinical management, as discordant responses between ctDNA and scans have been documented60, possibly due, in part, to tumour heterogeneity and the subclonal nature of metastatic breast cancer. For this method of surveillance to become clinically relevant, assay sensitivity, subclonal dynamics, and the emergence of resistance mutations need further study and development.
Treatment resistance and iterative drug choice
Despite the existence of effective treatments for early-stage and late-stage breast cancer, the majority of metastatic breast cancers will develop resistance. Many mechanisms of resistance exist, including those mediated by microenvironmental and metabolic factors, but the acquisition of mutations is an established mechanism in breast cancer61,62. The potential for ctDNA, accessible in plasma samples throughout most of the body, to contain subclonal mutations makes it an ideal tool for revealing somatic mutations in the metastatic setting. The surveillance of newly arising mutations provides a method for monitoring progression and drug resistance in real time. This approach could allow clinicians to adjust treatment courses and might improve patient outcome. Although many alterations responsible for therapeutic resistance have been identified in breast cancer, few studies have identified alternative treatment courses to overcome resistance based on these specific mutations. Currently, clinical trials are making use of ctDNA to address these needs (NCT03182634; NCT03778931; NCT02102165). For example, an early assessment of plasmaMATCH (NCT03182634) showed that the detection in ctDNA of targetable mutations could lead to effective therapy in pre-treated populations. In this study, mutations in AKT1 and ERBB2 were targeted with capivasertib and neratinib, respectively, leading to confirmed responses in four out of 18 and five out of 20 patients63 Although other arms of this study did not show similar responses, these results indicate the potential for mutation-guided treatment in late-stage, pre-treated breast cancer patients. Below, we will briefly discuss two prevalent mechanisms of resistance relevant to metastatic breast cancer: ESR1 mutations against hormone therapy, and various downstream mutations against PI3Kα inhibition.
ESR1 mutations
Resistance to endocrine therapy can evolve through multiple mechanisms, including the acquisition of mutations in ESR1, which encodes ERα. In 2013, Toy et al. and Robinson et al. were among the first to establish that ESR1 mutations constitute a major mechanism of resistance to endocrine therapy in patients with HR+ metastatic breast cancer64,65, occurring in approximately 30–40% of cases, particularly after treatment with aromatase inhibitors. Studies using ctDNA have shown that ESR1-mutant cells are rare in primary breast cancer but undergo subclonal expansion when tumours are exposed to endocrine therapy, further supporting their role in treatment resistance66,67,68,69,70. The utilisation of ctDNA in clinical studies has allowed clinicians to better understand metastatic disease and provide better options for standard of care therapy. The combined use of endocrine therapy and CDK4/6 inhibitors has recently been established as a standard of care for patients with HR+ metastatic breast cancer71,72,73. Results from the PALOMA-3 study revealed early ctDNA dynamics may provide a biomarker for CDK4/6 inhibitors and, as previously mentioned, may demonstrate divergent response of subclones to treatment58. Furthermore, ctDNA analysis from the PALOMA-3 trial revealed that treatment with fulvestrant alone and/or in combination with palbociclib led to the emergence of the ESR1 Y537S mutation, which confers constitutive activation of the ER (and, consequently, resistance to treatment)74. The PADA-1 trial, which evaluated the utility of ctDNA to monitor the onset of ESR1 mutations after treatment with a combination endocrine therapy and CDK4/6 inhibitor, found that the benefit is short-lived in metastatic breast cancer patients with the ESR1 mutation compared with those with wild-type tumours75. Finally, a combined analysis of the SoFEA and EFECT trials of patients with metastatic breast cancer who progressed on non-steroidal aromatase inhibitors showed that patients with ctDNA-detected ESR1 mutations showed increased overall survival and progression-free survival when treated with fulvestrant instead of exemestane76. This treatment strategy continues to be probed in the clinic through studies such as INTERACT (NCT04256941). Additionally, orally bioavailable selective ER degraders with similar mechanisms of action to fulvestrant are under development and have the potential to improve quality of life and outcome77,78.
PI3Kα inhibition
PIK3CA is the most commonly mutated gene in breast cancer and can be selected for during treatment. Mutations in this gene are often truncal driver mutations, with the same mutation present at the beginning of therapy and at relapse58. However, PIK3CA mutations can be dynamic throughout treatment, indicating that screening and rescreening using ctDNA could be important for treatments that target these mutations in particular79. In 2019, the PI3Kα inhibitor alpelisib was FDA approved in combination with hormone therapy to target HR+ PIK3CA mutant metastatic breast cancer80, concurrent with the approval of the Therascreen PIK3CA RGQ PCR kit as a companion diagnostic to detect mutations in tumours and the blood for patients conscripting onto alpelisib treatment. Unfortunately, a significant number of patients develop resistance to alpelisib. Loss of PTEN is one of the most well validated genetic resistance mechanisms that arises in response to PI3Kα inhibition81,82, but the ability to analyse ctDNA has been instrumental in identifying additional putative resistance mutations. A 2020 analysis of ctDNA pre- and post-progression on alpelisib in combination with aromatase inhibitors found that resistance mutations converged on genes downstream of the PI3K and mitogen-activated protein kinase (MAPK) pathways82. Therapies against many of these targets have already been developed but have yet to be extensively tested in this setting.
Thus, by using ctDNA for tumour surveillance in metastatic disease, the detection of resistance mutations might facilitate clinical decision making prior to clinical/radiographic progression while on therapy. This would ideally lead to an earlier change in therapy, with improved and personalised treatment outcomes for patients with metastatic breast cancer.
Conclusions
The detection and analysis of ctDNA from liquid biopsies has the potential to revolutionise clinical oncology. Methods for the use of ctDNA in early detection, patient stratification, therapy selection and response monitoring are currently being investigated, and provide the opportunity for a significant paradigm shift in the clinical setting. However, large-scale clinical trials to determine the clinical utility in tumour surveillance and to address previous limitations with sensitivity and duration of follow-up are still required. The effective application of ctDNA for late-stage disease monitoring, enrolment onto trials or off-label use of targeted therapies still requires an intimate knowledge of clonal dynamics, mutation function and drug efficacy. Despite these challenges, however, the potential for ctDNA to guide clinical decision making remains high, and the prospect of using liquid biopsy approaches in the provision of personalised care for patients with breast cancer continues to fuel laboratory research and future clinical trials.
References
Meisel, J. L., Venur, V. A., Gnant, M. & Carey, L. Evolution of targeted therapy in breast cancer: where precision medicine began. Am. Soc. Clin. Oncol. Educ. Book 38, 78–86 (2018).
Condorelli, R., Mosele, F., Verret, B., Bachelot, T., Bedard, P. L., Cortes, J. et al. Genomic alterations in breast cancer: level of evidence for actionability according to ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann. Oncol. 30, 365–373 (2019).
Overman, M. J., Modak, J., Kopetz, S., Murthy, R., Yao, J. C., Hicks, M. E. et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? J. Clin. Oncol. 31, 17–22 (2013).
Poulet, G., Massias, J. & Taly, V. Liquid biopsy: general concepts. Acta Cytol. 63, 449–455 (2019).
Chae, Y. K., Davis, A. A., Jain, S., Santa-Maria, C., Flaum, L., Beaubier, N. et al. Concordance of genomic alterations by next-generation sequencing in tumor tissue versus circulating tumor DNA in breast cancer. Mol. Cancer Ther. 16, 1412–1420 (2017).
Davis, A. A., Jacob, S., Gerratana, L., Shah, A. N., Wehbe, F., Katam, N. et al. Landscape of circulating tumour DNA in metastatic breast cancer. EBioMedicine 58, 102914 (2020).
Nishimura, F., Uno, N., Chiang, P. C., Kaku, N., Morinaga, Y., Hasegawa, H. et al. The effect of in vitro hemolysis on measurement of cell-free DNA. J. Appl. Lab. Med. 4, 235–240 (2019).
Sorber, L., Zwaenepoel, K., De Winne, K., Van Casteren, K., Augustus, E., Jacobs, J. et al. A multicenter study to assess EGFR mutational status in plasma: focus on an optimized workflow for liquid biopsy in a clinical setting. Cancers 10, 290 (2018).
Toro, P. V., Erlanger, B., Beaver, J. A., Cochran, R. L., VanDenBerg, D. A., Yakim, E. et al. Comparison of cell stabilizing blood collection tubes for circulating plasma tumor DNA. Clin. Biochem. 48, 993–998 (2015).
Toro, P. V., Erlanger, B., Beaver, J. A., Cochran, R. L., VanDenBerg, D. A., Yakim, E. et al. Comparison of cell stabilizing blood collection tubes for circulating plasma tumor DNA. Clin. Biochem. https://doi.org/10.1016/j.clinbiochem.2015.07.097 (2015).
Medford, A. J., Gillani, R. N. & Park, B. H. Detection of cancer DNA in early stage and metastatic breast cancer patients. Methods Mol. Biol. 1768, 209–227 (2018).
Sorenson, G. D., Pribish, D. M., Valone, F. H., Memoli, V. A., Bzik, D. J. & Yao, S. L. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol. Biomarkers Prev. 3, 67–71 (1994).
Hindson, C. M., Chevillet, J. R., Briggs, H. A., Gallichotte, E. N., Ruf, I. K., Hindson, B. J. et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 10, 1003–1005 (2013).
Miyazawa, H., Tanaka, T., Nagai, Y., Matsuoka, M., Huqun, Sutani, A. et al. Peptide nucleic acid-locked nucleic acid polymerase chain reaction clamp-based detection test for gefitinib-refractory T790M epidermal growth factor receptor mutation. Cancer Sci. 99, 595–600 (2008).
Wang, H., Jiang, J., Mostert, B., Sieuwerts, A., Martens, J. W., Sleijfer, S. et al. Allele-specific, non-extendable primer blocker PCR (AS-NEPB-PCR) for DNA mutation detection in cancer. J. Mol. Diagn. 15, 62–69 (2013).
Diehl, F., Li, M., He, Y., Kinzler, K. W., Vogelstein, B. & Dressman, D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat. Methods 3, 551–559 (2006).
Taniguchi, K., Uchida, J., Nishino, K., Kumagai, T., Okuyama, T., Okami, J. et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin. Cancer Res. 17, 7808–7815 (2011).
Hindson, B. J., Ness, K. D., Masquelier, D. A., Belgrader, P., Heredia, N. J., Makarewicz, A. J. et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610 (2011).
O’Leary, B., Hrebien, S., Beaney, M., Fribbens, C., Garcia-Murillas, I., Jiang, J. et al. Comparison of BEAMing and Droplet Digital PCR for Circulating Tumor DNA Analysis. Clin Chem 65, 1405–1413 (2019).
Taylor, S. C., Laperriere, G. & Germain, H. Droplet digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci. Rep. 7, 2409 (2017).
Chen, M. & Zhao, H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum. Genomics 13, 34 (2019).
El Achi, H., Khoury, J. D. & Loghavi, S. Liquid biopsy by next-generation sequencing: a multimodality test for management of cancer. Curr. Hematol. Malig. Rep. 14, 358–367 (2019).
Glenn, T. C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 11, 759–769 (2011).
Kinde, I., Wu, J., Papadopoulos, N., Kinzler, K. W. & Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl Acad. Sci. USA 108, 9530–9535 (2011).
Schmitt, M. W., Kennedy, S. R., Salk, J. J., Fox, E. J., Hiatt, J. B. & Loeb, L. A. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl Acad. Sci. USA 109, 14508–14513 (2012).
Phallen, J., Sausen, M., Adleff, V., Leal, A., Hruban, C., White, J. et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 9, eaan2415 (2017).
Newman, A. M., Lovejoy, A. F., Klass, D. M., Kurtz, D. M., Chabon, J. J., Scherer, F. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020).
Siu, A. L. & Force, U. S. P. S. T. Screening for breast cancer: U.S. preventive services task force recommendation statement. Ann. Intern. Med. 164, 279–296 (2016).
Agassi, R., Czeiger, D., Shaked, G., Avriel, A., Sheynin, J., Lavrenkov, K. et al. Measurement of circulating cell-free DNA levels by a simple fluorescent test in patients with breast cancer. Am. J. Clin. Pathol. 143, 18–24 (2015).
Gong, B., Xue, J., Yu, J., Li, H., Hu, H., Yen, H. et al. Cell-free DNA in blood is a potential diagnostic biomarker of breast cancer. Oncol. Lett. 3, 897–900 (2012).
Yu, D., Tong, Y., Guo, X., Feng, L., Jiang, Z., Ying, S. et al. Diagnostic value of concentration of circulating cell-free DNA in breast cancer: a meta-analysis. Front. Oncol. 9, 95 (2019).
Peled, M., Agassi, R., Czeiger, D., Ariad, S., Riff, R., Rosenthal, M. et al. Cell-free DNA concentration in patients with clinical or mammographic suspicion of breast cancer. Sci. Rep. 10, 14601 (2020).
Beaver, J. A., Jelovac, D., Balukrishna, S., Cochran, R., Croessmann, S., Zabransky, D. J. et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin. Cancer Res. 20, 2643–2650 (2014).
Bettegowda, C., Sausen, M., Leary, R. J., Kinde, I., Wang, Y., Agrawal, N. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra224 (2014).
Chan, K. C. A., Woo, J. K. S., King, A., Zee, B. C. Y., Lam, W. K. J., Chan, S. L. et al. Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N. Engl. J. Med. 377, 513–522 (2017).
Cohen, J. D., Li, L., Wang, Y., Thoburn, C., Afsari, B., Danilova, L. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
Lennon, A. M., Buchanan, A. H., Kinde, I., Warren, A., Honushefsky, A., Cohain, A. T. et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 369, eabb9601 (2020).
de Almeida, B. P., Apolonio, J. D., Binnie, A. & Castelo-Branco, P. Roadmap of DNA methylation in breast cancer identifies novel prognostic biomarkers. BMC Cancer 19, 219 (2019).
Xu, Z., Sandler, D. P. & Taylor, J. A. Blood DNA methylation and breast cancer: a prospective case-cohort analysis in the sister study. J. Natl Cancer Inst. 112, 87–94 (2020).
Shen, S. Y., Singhania, R., Fehringer, G., Chakravarthy, A., Roehrl, M. H. A., Chadwick, D. et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018).
Liu, M. C., Jamshidi, A., Venn, O., Fields, A. P., Maher, M. C., Cann, G. et al. Genome-wide cell-free DNA (cfDNA) methylation signatures and effect on tissue of origin (TOO) performance. J. Clin. Oncol. 37, 3049–3049 (2019).
Rothe, F., Silva, M. J., Venet, D., Campbell, C., Bradburry, I., Rouas, G. et al. Circulating tumor DNA in HER2-amplified breast cancer: a translational research substudy of the NeoALTTO Phase III Trial. Clin. Cancer Res. 25, 3581–3588 (2019).
Oshiro, C., Kagara, N., Naoi, Y., Shimoda, M., Shimomura, A., Maruyama, N. et al. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res. Treat. 150, 299–307 (2015).
Goh, J. Y., Feng, M., Wang, W., Oguz, G., Yatim, S., Lee, P. L. et al. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat. Med. 23, 1319–1330 (2017).
Widschwendter, M., Evans, I., Jones, A., Ghazali, S., Reisel, D., Ryan, A. et al. Methylation patterns in serum DNA for early identification of disseminated breast cancer. Genome Med. 9, 115 (2017).
Cavallone, L., Aguilar-Mahecha, A., Lafleur, J., Brousse, S., Aldamry, M., Roseshter, T. et al. Prognostic and predictive value of circulating tumor DNA during neoadjuvant chemotherapy for triple negative breast cancer. Sci. Rep. 10, 14704 (2020).
Riva, F., Bidard, F. C., Houy, A., Saliou, A., Madic, J., Rampanou, A. et al. Patient-specific circulating tumor DNA detection during neoadjuvant chemotherapy in triple-negative breast cancer. Clin. Chem. 63, 691–699 (2017).
McDonald, B. R., Contente-Cuomo, T., Sammut, S. J., Odenheimer-Bergman, A., Ernst, B., Perdigones, N. et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 11, eaax7392 (2019).
Cheng, L., Swartz, M. D., Zhao, H., Kapadia, A. S., Lai, D., Rowan, P. J. et al. Hazard of recurrence among women after primary breast cancer treatment-a 10-year follow-up using data from SEER-Medicare. Cancer Epidemiol. Biomarkers Prev. 21, 800–809 (2012).
Garcia-Murillas, I., Schiavon, G., Weigelt, B., Ng, C., Hrebien, S., Cutts, R. J. et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 7, 302ra133 (2015).
Olsson, E., Winter, C., George, A., Chen, Y., Howlin, J., Tang, M. H. et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol. Med. 7, 1034–1047 (2015).
Parsons, H. A., Rhoades, J., Reed, S. C., Gydush, G., Ram, P., Exman, P. et al. Sensitive detection of minimal residual disease in patients treated for early-stage breast cancer. Clin. Cancer Res. 26, 2556–2564 (2020).
Coombes, R. C., Page, K., Salari, R., Hastings, R. K., Armstrong, A., Ahmed, S. et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin. Cancer Res. 25, 4255–4263 (2019).
Dawson, S. J., Tsui, D. W., Murtaza, M., Biggs, H., Rueda, O. M., Chin, S. F. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013).
Kodahl, A. R., Ehmsen, S., Pallisgaard, N., Jylling, A. M. B., Jensen, J. D., Laenkholm, A. V. et al. Correlation between circulating cell-free PIK3CA tumor DNA levels and treatment response in patients with PIK3CA-mutated metastatic breast cancer. Mol. Oncol. 12, 925–935 (2018).
Liang, D. H., Ensor, J. E., Liu, Z. B., Patel, A., Patel, T. A., Chang, J. C. et al. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res. Treat. 155, 139–149 (2016).
O’Leary, B., Hrebien, S., Morden, J. P., Beaney, M., Fribbens, C., Huang, X. et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat. Commun. 9, 896 (2018).
Karthikeyan, S., Waters, I. G., Dennison, L., Chu, D., Donaldson, J., Shin, D. H. et al. Hierarchical tumor heterogeneity mediated by cell contact between distinct genetic subclones. J. Clin. Invest. 131, e143557 (2021).
Frenel, J. S., Carreira, S., Goodall, J., Roda, D., Perez-Lopez, R., Tunariu, N. et al. Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin. Cancer Res. 21, 4586–4596 (2015).
Housman, G., Byler, S., Heerboth, S., Lapinska, K., Longacre, M., Snyder, N. et al. Drug resistance in cancer: an overview. Cancers 6, 1769–1792 (2014).
Longley, D. B. & Johnston, P. G. Molecular mechanisms of drug resistance. J. Pathol. 205, 275–292 (2005).
Turner, N. C., Kingston, B., Kilburn, L. S., Kernaghan, S., Wardley, A. M., Macpherson, I. R. et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): a multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 21, 1296–1308 (2020).
Toy, W., Shen, Y., Won, H., Green, B., Sakr, R. A., Will, M. et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 45, 1439–1445 (2013).
Robinson, D. R., Wu, Y. M., Vats, P., Su, F., Lonigro, R. J., Cao, X. et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 45, 1446–1451 (2013).
Schiavon, G., Hrebien, S., Garcia-Murillas, I., Cutts, R. J., Pearson, A., Tarazona, N. et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci. Transl. Med. 7, 313ra182 (2015).
Chandarlapaty, S., Chen, D., He, W., Sung, P., Samoila, A., You, D. et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 Clinical Trial. JAMA Oncol. 2, 1310–1315 (2016).
Spoerke, J. M., Gendreau, S., Walter, K., Qiu, J., Wilson, T. R., Savage, H. et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat. Commun. 7, 11579 (2016).
Fribbens, C., O’Leary, B., Kilburn, L., Hrebien, S., Garcia-Murillas, I., Beaney, M. et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J. Clin. Oncol. 34, 2961–2968 (2016).
Wang, P., Bahreini, A., Gyanchandani, R., Lucas, P. C., Hartmaier, R. J., Watters, R. J. et al. Sensitive detection of mono- and polyclonal ESR1 mutations in primary tumors, metastatic lesions, and cell-free DNA of breast cancer patients. Clin. Cancer Res. 22, 1130–1137 (2016).
Li, J., Huo, X., Zhao, F., Ren, D., Ahmad, R., Yuan, X. et al. Association of cyclin-dependent kinases 4 and 6 inhibitors with survival in patients with hormone receptor-positive metastatic breast cancer: a systematic review and meta-analysis. JAMA Netw. Open 3, e2020312 (2020).
Sledge, G. W., Jr., Toi, M., Neven, P., Sohn, J., Inoue, K., Pivot, X. et al. The effect of abemaciclib plus fulvestrant on overall survival in hormone receptor-positive, ERBB2-negative breast cancer that progressed on endocrine therapy-MONARCH 2: a randomized clinical trial. JAMA Oncol. https://doi.org/10.1001/jamaoncol.2019.4782 (2019).
Turner, N. C., Slamon, D. J., Ro, J., Bondarenko, I., Im, S. A., Masuda, N. et al. Overall survival with palbociclib and fulvestrant in advanced breast cancer. N. Engl. J. Med. 379, 1926–1936 (2018).
O’Leary, B., Cutts, R. J., Liu, Y., Hrebien, S., Huang, X., Fenwick, K. et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 8, 1390–1403 (2018).
Bidard, F. C., Callens, C., Dalenc, F., Pistilli, B., Rouge, T. D. L. M., Clatot, F. et al. Prognostic impact of ESR1 mutations in ER+ HER2- MBC patients prior treated with first line AI and palbociclib: an exploratory analysis of the PADA-1 trial. J. Clin. Oncol. 38, 1010 (2020).
Turner, N. C., Swift, C., Kilburn, L., Fribbens, C., Beaney, M., Garcia-Murillas, I. et al. ESR1 mutations and overall survival on fulvestrant versus exemestane in advanced hormone receptor-positive breast cancer: a combined analysis of the Phase III SoFEA and EFECT Trials. Clin. Cancer Res. 26, 5172–5177 (2020).
Bardia, A., Aftimos, P., Bihani, T., Anderson-Villaluz, A. T., Jung, J., Conlan, M. G. et al. EMERALD: phase III trial of elacestrant (RAD1901) vs endocrine therapy for previously treated ER+ advanced breast cancer. Fut. Oncol. 15, 3209–3218 (2019).
Kaklamani, V., Bardia, A., Wilks, S., Weise, A., Richards, D., Harb, W. et al. Final analysis of phase 1 study of elacestrant (RAD1901), a novel selective estrogen receptor degrader (SERD), in estrogen receptor positive (ER+), human epidermal growth factor receptor 2 negative (HER2-) advanced breast cancer. Cancer Res. 80, PD7–PD07 (2020).
Higgins, M. J., Jelovac, D., Barnathan, E., Blair, B., Slater, S., Powers, P. et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin. Cancer Res. 18, 3462–3469 (2012).
Andre, F., Ciruelos, E., Rubovszky, G., Campone, M., Loibl, S., Rugo, H. S. et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 380, 1929–1940 (2019).
Juric, D., Castel, P., Griffith, M., Griffith, O. L., Won, H. H., Ellis, H. et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature 518, 240–244 (2015).
Razavi, P. Alterations in PTEN and ESR1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nat. Cancer 1, 382–393 (2020).
Acknowledgements
The authors thank members of the Park Lab for their thoughtful comments.
Author information
Authors and Affiliations
Contributions
B.A.D., S.C. and B.H.P. conceived the design and wrote the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Data availability
Not applicable.
Competing interests
B.H.P. is a paid consultant for Jackson Labs, Casdin Capital, Pathovax, Sermonix and is a paid scientific advisory board member for Celcuity Inc. Under separate licensing agreements between Horizon Discovery, Ltd and The Johns Hopkins University, S.C. and B.H.P. are entitled to a share of royalties received by the University on sales of products. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. B.A.D. declares no potential conflicts of interest.
Funding information
This work was supported by: The Breast Cancer Research Foundation, Komen Foundation, NIH CA214494, CA194024 (B.H.P.). We would also like to thank and acknowledge the support of The Canney Foundation, the Marcie and Ellen Foundation, Amy and Barry Baker, Donna and John Hall and the Vanderbilt-Ingram Cancer Center support grant (NIH CA068485) and Breast Cancer SPORE (NIH CA098131).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Davidson, B.A., Croessmann, S. & Park, B.H. The breast is yet to come: current and future utility of circulating tumour DNA in breast cancer. Br J Cancer 125, 780–788 (2021). https://doi.org/10.1038/s41416-021-01422-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01422-w