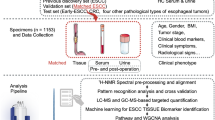
Abstract
Background
Oesophageal cancer (EC) ranks high in both morbidity and mortality. A non-invasive and high-sensitivity diagnostic approach is necessary to improve the prognosis of EC patients.
Methods
A total of 525 serum samples were subjected to lipidomic analysis. We combined serum lipidomics and machine-learning algorithms to select important metabolite features for the detection of oesophageal squamous cell carcinoma (ESCC), the major subtype of EC in developing countries. A diagnostic model using a panel of selected features was developed and evaluated. Integrative analyses of tissue transcriptome and serum lipidome were conducted to reveal the underlying mechanism of lipid dysregulation.
Results
Our optimised diagnostic model with a panel of 12 lipid biomarkers together with age and gender reaches a sensitivity of 90.7%, 91.3% and 90.7% and an area under receiver-operating characteristic curve of 0.958, 0.966 and 0.818 in detecting ESCC for the training cohort, validation cohort and independent validation cohort, respectively. Integrative analysis revealed matched variation trend of genes encoding key enzymes in lipid metabolism.
Conclusions
We have identified a panel of 12 lipid biomarkers for diagnostic modelling and potential mechanisms of lipid dysregulation in the serum of ESCC patients. This is a reliable, rapid and non-invasive tumour-diagnostic approach for clinical application.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Domper Arnal, M. J., Ferrandez Arenas, A. & Lanas Arbeloa, A. Esophageal cancer: risk factors, screening and endoscopic treatment in Western and Eastern countries. World J. Gastroenterol. 21, 7933–7943 (2015).
Rustgi, A. K. & El-Serag, H. B. Esophageal carcinoma. N. Engl. J. Med. 371, 2499–2509 (2014).
Lagergren, J., Smyth, E., Cunningham, D. & Lagergren, P. Oesophageal cancer. Lancet 390, 2383–2396 (2017).
Lao-Sirieix, P. & Fitzgerald, R. C. Screening for oesophageal cancer. Nat. Rev. Clin. Oncol. 9, 278–287 (2012).
Liu, M., He, Z., Guo, C., Xu, R., Li, F., Ning, T. et al. Effectiveness of intensive endoscopic screening for esophageal cancer in China: a community-based study. Am. J. Epidemiol. 188, 776–784 (2019).
Chen, X. X., Zhong, Q., Liu, Y., Yan, S. M., Chen, Z. H., Jin, S. Z. et al. Genomic comparison of esophageal squamous cell carcinoma and its precursor lesions by multi-region whole-exome sequencing. Nat. Commun. 8, 524 (2017).
Cheng, C., Geng, F., Cheng, X. & Guo, D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 38, 27 (2018).
Snaebjornsson, M. T., Janaki-Raman, S. & Schulze, A. Greasing the wheels of the cancer machine: the role of lipid metabolism in cancer. Cell Metab. 31, 62–76 (2020).
Perrotti, F., Rosa, C., Cicalini, I., Sacchetta, P., Del Boccio, P., Genovesi, D. et al. Advances in lipidomics for cancer biomarkers discovery. Int. J. Mol. Sci. 17, https://doi.org/10.3390/ijms17121992 (2016).
Costello, E. A metabolomics-based biomarker signature discriminates pancreatic cancer from chronic pancreatitis. Gut 67, 2–3 (2018).
Li, J., Ren, S., Piao, H. L., Wang, F., Yin, P., Xu, C. et al. Integration of lipidomics and transcriptomics unravels aberrant lipid metabolism and defines cholesteryl oleate as potential biomarker of prostate cancer. Sci. Rep. 6, 20984 (2016).
Sun, C., Li, T., Song, X., Huang, L., Zang, Q., Xu, J. et al. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc. Natl Acad. Sci. USA 116, 52–57 (2019).
Chu, L. Y., Peng, Y. H., Weng, X. F., Xie, J. J. & Xu, Y. W. Blood-based biomarkers for early detection of esophageal squamous cell carcinoma. World J. Gastroenterol. 26, 1708–1725 (2020).
Mir, S. A., Rajagopalan, P., Jain, A. P., Khan, A. A., Datta, K. K., Mohan, S. V. et al. LC-MS-based serum metabolomic analysis reveals dysregulation of phosphatidylcholines in esophageal squamous cell carcinoma. J. Proteom. 127, 96–102 (2015).
Zhu, Y., Zhang, H., Chen, N., Hao, J., Jin, H. & Ma, X. Diagnostic value of various liquid biopsy methods for pancreatic cancer: a systematic review and meta-analysis. Medicine 99, e18581 (2020).
Mayerle, J., Kalthoff, H., Reszka, R., Kamlage, B., Peter, E., Schniewind, B. et al. Metabolic biomarker signature to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Gut 67, 128–137 (2018).
Merker, J. D., Oxnard, G. R., Compton, C., Diehn, M., Hurley, P., Lazar, A. J. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 36, 1631–1641 (2018).
Balasenthil, S., Huang, Y., Liu, S., Marsh, T., Chen, J., Stass, S. A. et al. A plasma biomarker panel to identify surgically resectable early-stage pancreatic cancer. J. Natl. Cancer Inst. 109, https://doi.org/10.1093/jnci/djw341 (2017).
Huang, L., Wang, L., Hu, X., Chen, S., Tao, Y., Su, H. et al. Machine learning of serum metabolic patterns encodes early-stage lung adenocarcinoma. Nat. Commun. 11, 3556 (2020).
Mathe, E. A., Patterson, A. D., Haznadar, M., Manna, S. K., Krausz, K. W., Bowman, E. D. et al. Noninvasive urinary metabolomic profiling identifies diagnostic and prognostic markers in lung cancer. Cancer Res. 74, 3259–3270 (2014).
Maros, M. E., Capper, D., Jones, D. T. W., Hovestadt, V., von Deimling, A., Pfister, S. M. et al. Machine learning workflows to estimate class probabilities for precision cancer diagnostics on DNA methylation microarray data. Nat. Protoc. 15, 479–512 (2020).
Li, R., Li, Y., Kristiansen, K. & Wang, J. SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714 (2008).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zhou, J. & Yin, Y. Use of liquid chromatography-mass spectrometry-based metabolomics to identify biomarkers of tuberculosis. Methods Mol. Biol. 1859, 241–251 (2019).
Rohrig, F. & Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749 (2016).
Yoon, S., Lee, M. Y., Park, S. W., Moon, J. S., Koh, Y. K., Ahn, Y. H. et al. Up-regulation of acetyl-CoA carboxylase alpha and fatty acid synthase by human epidermal growth factor receptor 2 at the translational level in breast cancer cells. J. Biol. Chem. 282, 26122–26131 (2007).
Nath, A., Li, I., Roberts, L. R. & Chan, C. Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma. Sci. Rep. 5, 14752 (2015).
Yue, S., Li, J., Lee, S. Y., Lee, H. J., Shao, T., Song, B. et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 19, 393–406 (2014).
Pascual, G., Avgustinova, A., Mejetta, S., Martin, M., Castellanos, A., Attolini, C. S. et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 541, 41–45 (2017).
Wu, H., Yu, J., Li, Y., Hou, Q., Zhou, R., Zhang, N. et al. Single-cell RNA sequencing reveals diverse intratumoral heterogeneities and gene signatures of two types of esophageal cancers. Cancer Lett. 438, 133–143 (2018).
Li, C., Wang, Q., Ma, J., Shi, S., Chen, X., Yang, H. et al. Integrative pathway analysis of genes and metabolites reveals metabolism abnormal subpathway regions and modules in esophageal squamous cell carcinoma. Molecules 22, https://doi.org/10.3390/molecules22101599 (2017).
Das, N., Topalovic, M. & Janssens, W. Artificial intelligence in diagnosis of obstructive lung disease: current status and future potential. Curr. Opin. Pulm. Med. 24, 117–123 (2018).
Zhou, L. Q., Wang, J. Y., Yu, S. Y., Wu, G. G., Wei, Q., Deng, Y. B. et al. Artificial intelligence in medical imaging of the liver. World J. Gastroenterol. 25, 672–682 (2019).
Acknowledgements
We thank Dr. Z. Zhou for the analysis of transcriptomic data.
Author information
Authors and Affiliations
Contributions
Y. Yin. and Y.S. designed this study. Y. Yuan, H.S., G.W., J.Z. and R.P. conducted sample preparation and LC-MS analysis. Z.Z., L.X. and Y.S. collected clinical samples and clinical information and generated transcriptomic data. Y. Yuan and G.W. conducted data analysis and interpreted the data with J.Z., R.P. and L.J. Y. Yuan, Y.S. and Y. Yin wrote the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was performed in accordance with the Declaration of Helsinki and was conducted after obtaining approval from the Independent Ethics Committee at the National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (No. NCC2596). Informed consent was obtained from all patients or healthy controls. All methods were performed in accordance with relevant guidelines and regulations.
Consent to publish
Not applicable.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Funding information
This work was supported by grants to Y. Yin, including the National Key Research and Development Programme of China (2016YFA0500302 to Y. Yin), the National Natural Scientific Foundation of China (31420103905, 81321003, 81430056 and 81372491 to Y. Yin), the Beijing Natural Science Foundation (Key grant 7161007 to Y. Yin), the Shu Fan Education and Research Foundation (to Y. Yin), and Lam Chung Nin Foundation for Systems Biomedicine (to Y. Yin).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yuan, Y., Zhao, Z., Xue, L. et al. Identification of diagnostic markers and lipid dysregulation in oesophageal squamous cell carcinoma through lipidomic analysis and machine learning. Br J Cancer 125, 351–357 (2021). https://doi.org/10.1038/s41416-021-01395-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-021-01395-w
This article is cited by
-
Prediction of positive pulmonary nodules based on machine learning algorithm combined with central carbon metabolism data
Journal of Cancer Research and Clinical Oncology (2024)
-
Data-driven decision-making for precision diagnosis of digestive diseases
BioMedical Engineering OnLine (2023)
-
Novel research and future prospects of artificial intelligence in cancer diagnosis and treatment
Journal of Hematology & Oncology (2023)