Abstract
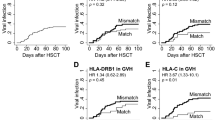
The “human leukocyte antigen (HLA) supertype” is a functional classification of HLA alleles, which was defined by structural features and peptide specificities, and has been reportedly associated with the clinical outcomes of viral infections and autoimmune diseases. Although the disparity in each HLA locus was reported to have no clinical significance in single-unit cord blood transplantation (sCBT), the clinical significance of the HLA supertype in sCBT remains unknown. Therefore, we retrospectively analyzed clinical data of 1603 patients who received sCBT in eight institutes in Japan between 2000 and 2017. Each HLA allele was categorized into 19 supertypes, and the prognostic effect of disparities was then assessed. An HLA-B supertype mismatch was identified as a poor prognostic factor (PFS: hazard ratio [HR] = 1.23, p = 0.00044) and was associated with a higher cumulative incidence (CI) of relapse (HR = 1.24, p = 0.013). However, an HLA-B supertype mismatch was not associated with the CI of acute and chronic graft-versus-host-disease. The multivariate analysis for relapse and PFS showed the significance of an HLA-B supertype mismatch independent of allelic mismatches, and other previously reported prognostic factors. HLA-B supertype-matched grafts should be selected in sCBT.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Eapen M, Rubinstein P, Zhang M-J, Stevens C, Kurtzberg J, Scaradavou A, et al. Outcomes of transplantation of unrelated donor umbilical cord blood and bone marrow in children with acute leukaemia: a comparison study. Lancet. 2007;369:1947–54.
Barker JN, Byam C, Scaradavou A. How I treat: the selection and acquisition of unrelated cord blood grafts. Blood. 2011;117:2332–9.
Kawase T, Morishima Y, Matsuo K, Kashiwase K, Inoko H, Saji H, et al. High-risk HLA allele mismatch combinations responsible for severe acute graft-versus-host disease and implication for its molecular mechanism. Blood. 2007;110:2235–41.
Morishima S, Kashiwase K, Matsuo K, Azuma F, Yabe T, Sato-Otsubo A, et al. High-risk HLA alleles for severe acute graft-versus-host disease and mortality in unrelated donor bone marrow transplantation. Haematologica. 2016;101:491–8.
Koyama M, Hill GR. Alloantigen presentation and graft-versus-host disease: fuel for the fire. Blood. 2016;127:2963–70.
Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1–15.
Lund O, Nielsen M, Kesmir C, Petersen AG, Lundegaard C, Worning P, et al. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics. 2004;55:797–810.
Harjanto S, Ng LFP, Tong JC. Clustering HLA class I superfamilies using structural interaction patterns. PLoS One 2014; 9. e86655. https://doi.org/10.1371/journal.pone.0086655.
Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–12.
Tong JC, Tan TW, Ranganathan S. In silico grouping of peptide/HLA class I complexes using structural interaction characteristics. Bioinformatics. 2007;23:177–83.
Doytchinova IA, Guan P, Flower DR. Identifiying human MHC supertypes using bioinformatic methods. J Immunol. 2004;172:4314–23.
Richardson J, Reyburn HT, Luque I, Valés‐Gómez M, Strominger JL. Definition of polymorphic residues on killer Ig‐like receptor proteins which contribute to the HLA‐C binding site. Eur J Immunol. 2000;30:1480–5.
Doytchinova IA, Flower DR. In silico identification of supertypes for class II MHCs. J Immunol. 2005;174:7085–95.
Trachtenberg E, Korber B, Sollars C, Kepler TB, Hraber PT, Hayes E, et al. Advantage of rare HLA supertype in HIV disease progression. Nat Med. 2003;9:928–35.
Deutekom HWM, van, Hoof I, Bontrop RE, Kesmir C. A comparative analysis of viral peptides presented by contemporary human and chimpanzee MHC class I molecules. J Immunol. 2011;187:5995–6001.
Lazaryan A, Song W, Lobashevsky E, Tang J, Shrestha S, Zhang K, et al. Human leukocyte antigen class I supertypes and HIV-1 control in African Americans. J Virol. 2010;84:2610–7.
Matsueda S, Takedatsu H, Sasada T, Azuma K, Ishihara Y, Komohara Y, et al. New peptide vaccine candidates for epithelial cancer patients With HLA-A3 supertype alleles. J Immunother. 2007;30:274–81.
Matsueda S, Takedatsu H, Yao A, Tanaka M, Noguchi M, Itoh K, et al. Identification of peptide vaccine candidates for prostate cancer patients with HLA-A3 supertype alleles. Clin Cancer Res. 2005;11:6933–43.
Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359:582–7.
Karosiene E, Lundegaard C, Lund O, Nielsen M. NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics. 2012;64:177–86.
Thomsen M, Lundegaard C, Buus S, Lund O, Nielsen M. MHCcluster, a method for functional clustering of MHC molecules. Immunogenetics. 2013;65:655–65.
Kalita P, Padhi AK, Zhang KYJ, Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Micro Pathog. 2020;145:104236.
Lazaryan A, Wang T, Spellman SR, Wang H-L, Pidala J, Nishihori T, et al. Human leukocyte antigen supertype matching after myeloablative hematopoietic cell transplantation with 7/8 matched unrelated donor allografts: a report from the Center for International Blood and Marrow Transplant Research. Haematologica. 2016;101:1267–74.
Sorror ML, Maris MB, Storb R, Baron F, Sandmaier BM, Maloney DG, et al. Hematopoietic cell transplantation (HCT)-specific comorbidity index: a new tool for risk assessment before allogeneic HCT. Blood. 2005;106:2912–9.
Armand P, Kim HT, Logan BR, Wang Z, Alyea EP, Kalaycio ME, et al. Validation and refinement of the Disease Risk Index for allogeneic stem cell transplantation. Blood. 2014;123:3664–71.
Giralt S, Ballen K, Rizzo D, Bacigalupo A, Horowitz M, Pasquini M, et al. Reduced-intensity conditioning regimen workshop: defining the dose spectrum. report of a workshop convened by the Center for International Blood and Marrow Transplant Research. Biol Blood Marrow Traspl. 2009;15:367–9.
Bacigalupo A, Ballen K, Rizzo D, Giralt S, Lazarus H, Ho V, et al. Defining the intensity of conditioning regimens: working definitions. Biol Blood Marrow Transpl. 2009;15:1628–33.
Gaballa A, Clave E, Uhlin M, Toubert A, Arruda LCM. Evaluating thymic function after human hematopoietic stem cell transplantation in the personalized medicine era. Front Immunol. 2020;11:1341.
Clave E, Lisini D, Douay C, Giorgiani G, Busson M, Zecca M, et al. Thymic function recovery after unrelated donor cord blood or T-cell depleted HLA-haploidentical stem cell transplantation correlates with leukemia relapse. Front Immunol. 2013;4:54.
Song ES, Linsk R, Olson CA, McMillan M, Goodenow RS. Allospecific cytotoxic T lymphocytes recognize an H-2 peptide in the context of a murine major histocompatibility complex class I molecule. Proc Natl Acad Sci. 1988;85:1927–31.
Kievits F, Ivanyi P. A subpopulation of mouse cytotoxic T lymphocytes recognizes allogeneic H-2 class I antigens in the context of other H-2 class I molecules. J Exp Med. 1991;174:15–19.
Krenger W, Holländer GA. The immunopathology of thymic GVHD. Semin Immunopathol. 2008;30:439.
Godthelp BC, van Tol MJ, Vossen JM, van Den Elsen PJ. T-cell immune reconstitution in pediatric leukemia patients after allogeneic bone marrow transplantation with T-cell–depleted or unmanipulated grafts: evaluation of overall and antigen-specific T-cell repertoires. Blood. 1999;94:4358–69.
Eyrich M, Croner T, Leiler C, Lang P, Bader P, Klingebiel T, et al. Distinct contributions of CD4+ and CD8+naive and memory T-cell subsets to overall T-cell–receptor repertoire complexity following transplantation of T-cell–depleted CD34-selected hematopoietic progenitor cells from unrelated donors. Blood. 2002;100:1915–8.
Roux E, Dumont-Girard F, Starobinski M, Siegrist CA, Helg C, Chapuis B, et al. Recovery of immune reactivity after T-cell-depleted bone marrow transplantation depends on thymic activity. Blood. 2000;96:2299–303.
Touma W, Brunstein CG, Cao Q, Miller JS, Curtsinger J, Verneris MR, et al. Dendritic cell recovery impacts outcomes after umbilical cord blood and sibling donor transplantation for hematologic malignancies. Biol Blood Marrow Transpl. 2017;23:1925–31.
Reddy P, Maeda Y, Liu C, Krijanovski OI, Korngold R, Ferrara JLM. A crucial role for antigen-presenting cells and alloantigen expression in graft-versus-leukemia responses. Nat Med. 2005;11:1244–9.
Prugnolle F, Manica A, Charpentier M, Guégan JF, Guernier V, Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Curr Biol. 2005;15:1022–7.
Francisco R, dos S, Buhler S, Nunes JM, Bitarello BD, França GS, et al. HLA supertype variation across populations: new insights into the role of natural selection in the evolution of HLA-A and HLA-B polymorphisms. Immunogenetics. 2015;67:651–63.
Lu Z, Chen H, Jiao X, Wang Y, Wu L, Sun H, et al. Germline HLA-B evolutionary divergence influences the efficacy of immune checkpoint blockade therapy in gastrointestinal cancer. Genome Med. 2021;13:175.
Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–75.
Proietto, Dommelen AI, van S, Zhou P, Rizzitelli A, D’Amico A, et al. Dendritic cells in the thymus contribute to T-regulatory cell induction. Proc Natl Acad Sci. 2008;105:19869–74.
Teshima T, Reddy P, Liu C, Williams D, Cooke KR, Ferrara JLM. Impaired thymic negative selection causes autoimmune graft-versus-host disease. Blood. 2003;102:429–35.
Wu T, Young JS, Johnston H, Ni X, Deng R, Racine J, et al. Thymic damage, impaired negative selection, and development of chronic graft-versus-host disease caused by donor CD4+ and CD8+ T cells. J Immunol. 2013;191:488–99.
Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–82.
Jiménez M, Martínez C, Ercilla G, Carreras E, Urbano-Ispízua Á, Aymerich M, et al. Reduced-intensity conditioning regimen preserves thymic function in the early period after hematopoietic stem cell transplantation. Exp Hematol. 2005;33:1240–8.
Chen X, Hale GA, Barfield R, Benaim E, Leung WH, Knowles J, et al. Rapid immune reconstitution after a reduced‐intensity conditioning regimen and a CD3‐depleted haploidentical stem cell graft for paediatric refractory haematological malignancies. Br J Haematol. 2006;135:524–32.
Castermans E, Hannon M, Dutrieux J, Humblet-Baron S, Seidel L, Cheynier R, et al. Thymic recovery after allogeneic hematopoietic cell transplantation with non-myeloablative conditioning is limited to patients younger than 60 years of age. Haematologica. 2011;96:298–306.
Palmer DB. The effect of age on thymic function. Front Immunol. 2013;4:316.
Terakura S, Wake A, Inamoto Y, Murata M, Sakai R, Yamaguchi T, et al. Exploratory research for optimal GvHD prophylaxis after single unit CBT in adults: short-term methotrexate reduced the incidence of severe GvHD more than mycophenolate mofetil. Bone Marrow Transpl. 2017;52:423–30.
Acknowledgements
We thank the patients who participated in this study and the medical and nursing staff for providing patient care. We also thank Eriko Makizumi Kabayama and Miyuki Yoshikawa for their assistance with the data collection.
Author information
Authors and Affiliations
Contributions
TS, NU, KM, SM, and KK designed experiments and wrote the manuscript. TS and NU collected the clinical data. TS, NU, KM, and KK analyzed the data. YO, TE, YM, GY, KY, YK, KN, HI, TK, RO, TM, ST, and KA reviewed the data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sugio, T., Uchida, N., Miyawaki, K. et al. Prognostic impact of HLA supertype mismatch in single-unit cord blood transplantation. Bone Marrow Transplant 59, 466–472 (2024). https://doi.org/10.1038/s41409-023-02183-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41409-023-02183-1