Abstract
Caucasian Americans (CA) compared with African Americans (AA) have a twofold increased incidence of multiple myeloma (MM) and have an earlier age of diagnosis. However, there is sparse information regarding underlying biological differences across racial/ethnic groups. We characterized genetic alterations using a targeted next-generation sequencing assay called myTYPE, developed at MSKCC, allowing capture of somatic mutations, IgH translocations, gains/losses, and hyperdiploidy. Samples were obtained from the NIH Plasma Cell Dyscrasia Racial Disparity Cohort. In total, 68 patient samples were successfully sequenced and manually curated based on well-established databases. Of the 68 patient samples (47 CA, 21 AA), 84% had at least one type of genomic alteration. Importantly, the IgH translocation, t(11;14), was observed more frequently in the AA group (0 vs. 29%, p = 0.001). Known oncogenic somatic non-synonymous mutations were found in 18 genes and indels in 2 genes. KRAS mutations were the most common mutation found in 16% of patients followed by NRAS and BRAF mutations. TP53 somatic mutations appeared to be more common in CA but lacked significance. This proof-of-principle study indicates the presence of varying underlying tumor biology between racial groups and supports the need of future prospective trials to capture these molecular characteristics.
Similar content being viewed by others
Introduction
Despite advancements in the understanding and treatment of multiple myeloma (MM), a racial disparity in clinical presentation and outcomes remain. Compared with Caucasian Americans (CA), African Americans (AA) matched for socioeconomics, age, and gender have a twofold increased incidence of MM, have an earlier average age at diagnosis by 5–10 years, and have gained less benefit from the advent of novel agents in the last decade1,2. These differences have not been shown to be attributable to disparities in access to medical care. In addition, over the past decade, improvements in survival with the introduction of proteasome inhibitors and immunomodulatory agents is predominantly observed in CA. Costa et al.3 observed improvements in 10-year relative survival rates (RSRs) in all racial groups < 65 years of age and no improvements for either racial group over 75 years of age. In patients between the ages of 65 and 74 years, CA had an improvement in 10-year RSRs but AA did not. Moreover, although it has been noted that AA have an increased myeloma-related mortality rate, this is in fact a reflection of the increased incidence of MM in AA rather than worse prognosis. In a pivotal study of 30,000 patients, the authors concluded that AA appear to have a better prognosis compared with CA4.
The variation in clinical course suggests an underlying molecular heterogeneity between races. Despite the increased frequency of MM among AA, most of the known molecular data and association with clinical outcomes, including traditional fluorescence in situ hybridization (FISH)/cytogenetics and newer NGS methods have been derived from CA cohorts5,6,7,8. At this time there is no single unifying genetic or genomic alteration known to cause MM but there are multiple alterations frequently identified. Approximately half of MM genomes are hyperdiploid (gain of an additional odd numbered chromosomes)9,10. Most of the non-hyperdiploid MM cases harbor a translocation involving the immunoglobulin heavy-chain (IgH) gene located on chromosome 149,10. These genetic lesions are thought to be primary events, as they are also found in the precursor state, monoclonal gammopathy of undetermined significance (MGUS)11. In ~10% of cases, both aberrations co-occur12,13. In general, hyperdiploid MM is associated with an improved prognosis compared with MM cases with an IgH translocation, except for the cyclin D translocations (t(6;14) and t(11;14)), which are considered neutral14,15. The five most frequent translocations in descending order are t(11;14), t(4;14), t(14;16), t(14;20), and t(6;14)15,16. Based on karyotyping and interphase FISH, t(4;14), t(14;16), and t(14;20) have been identified as high-risk primary genetic events, along with the secondary/tertiary events of deletion 17p, deletion 1p32, and 1q gains9,14. The genetic heterogeneity of myeloma is reflected in the variety of genetic hits including secondary translocations, copy number variants (CNVs), and somatic oncogenic mutations17.
To improve our understanding of the underlying biological mechanisms of the racial disparity in patients with MM, this study used a targeted NGS assay termed myTYPE developed at Memorial Sloan Kettering Cancer Center. myTYPE was specifically developed to target genomic aberrations known to occur in patients with MM18,19. The myTYPE assay is designed to capture known IgH translocations, hyperdiploidy, CNVs, and somatic mutations in 120 frequently mutated genes in MM. Using this specific assay we investigated the differences in somatic mutations, translocations, and chromosomal gains/losses between CA and AA MM patients.
Methods
Patients and techinical assays
Bone marrow clot sections were obtained from the National Institutes of Health Plasma Cell Dyscrasia Racial Disparity Cohort. A total of 91 pretreatment baseline samples from patients with newly diagnosed MM (NDMM) underwent DNA extraction, 81 samples met DNA quality control (QC) and purity criteria, and underwent NGS library preparation. Of these, 68 (47 CA, 21 AA) patient samples passed all QC measures for sequencing. In the myTYPE assay, baits were designed to capture the entire IgH locus (where the majority of the canonical chromosome 14 breakpoints occur) and the partner chromosome, genome-wide single-nucleotide polymorphisms for hyperdiploidy, and other CNVs as well as exons of 120 frequently mutated genes in MM. With this design, myTYPE detects the IGH translocations and the partner chromosome regardless of which the partner is. In addition, myTYPE detects hyperdiploidy, arm-level chromosomal gains and losses, as well as the most common and relevant somatic mutations. The target regions from bone marrow clot section samples were amplified and then sequenced using 126 base-paired end reads using Illumina HiSeq with a mean target depth of 413.5×. Data were analyzed using validated bioinformatic algorithms (Supplementary Methods). Race was determined from patient self-reporting. Fisher’s exact test was used to calculate two-tailed p-values for differences between CA and AA groups. The Bonferroni method was used to correct for multiple testing and control the family-wise error rate at <0.05. This resulted in a significance threshold of p < 0.0015 for each comparison.
Results
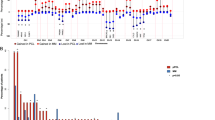
Of the 68 patient baseline samples sequenced (47 CA, 21 AA), 57 patients (87%) had at least one genomic alteration (i.e., hyperdiploidy, translocations, chromosomal gains/losses, indels, or somatic non-synonymous mutations). Of these, 20 (95%) were from AA and 37 (79%) from CA patient samples (Figs. 1 and 2). Putative oncogenic mutations and indels (insertions and deletions) were observed in 19 oncogenic genes (Table 1). KRAS mutations were most common, identified in 16% of patients (13% CA, 24% AA). NRAS (4% CA, 10% AA) and BRAF (2% CA, 14% AA) mutations were the second most common. BRAF mutations were all V600E non-synonymous mutations and statistically trended to be more common in AA (p = 0.08). TP53 somatic mutations also appeared to be more common in CA (6%) than AA (0%); however, this difference did not reach statistical significance. Oncogenic mutations were also observed in FAM46C, EGR1, PTPN11, ATM, CCND1, DIS3, FAT1, FGFR3, HIST1H1E, KDM6A, LTB, MAX, MYC, and SP140. Indels (frameshift and in-frame variants) were identified in FAM46C and CYLD. In 11 of the patient samples, no genomic alterations were observed.
Genetic alterations in African American (AA) patient samples. Sixteen of 21 AA patients myeloma samples had either an amplificant or deletion, IgH translocation, insertion/deletion, or oncogenic missense mutation. KRAS mutations were the most frequently mutated gene. Compared with Caucasian Americans, AA were enriched for t(11;14)
Genetic alterations in Caucasian American (CA) patient samples. Thirty-two of 47 CA patients’ myeloma samples had either an amplificant or deletion, IgH translocation, insertion/deletion, or oncogenic missense mutation. KRAS mutations were the most frequently mutated gene. Compared with African Americans, CA had an increased frequency of TP53 oncogenic mutations but did not reach statistical significance
Four different translocations involving IgH were identified in patients (Table 2). The t(11;14) occurred significantly more frequently in the AA group (29% vs. 0, p = 0.0005). This difference remained significant when adjusting for multiple testing. The incidence of t(4;14), t(8;14), and t(14;16) were not significantly different between the CA and AA groups. Chromosomal gains and losses were identified in many patients and, as expected, approximately half the patients had hyperdiploid myeloma (Table 2). Moreover, del(1p) was observed more commonly in the AA population (0 vs. 14%, p = 0.037) as well as amp(1q) (13% vs. 33%, p = 0.091); however, neither remained significant when adjusting for multiple testing. There were no other significant differences between racial groups including patients with hyperdiploid genomes, del(17p), or del(13q).
Given the striking observation that t(11;14) was more common in AA patient samples, IgH translocation data were downloaded from the Multiple Myeloma Research Foundation (MMRF) CoMMpass (Relating Clinical Outcomes in MM to Personal Assessment of Genetic Profile) Study (research.themmrf.org) to analyze and determine whether racial differences in t(11;14) were observed in a larger cohort of patients20. To the best of our knowledge, racial differences in translocations in this cohort had not been previously reported. Based on validated racial classification previously reported, 658 samples were evaluable for chromosome 14 translocations (112 AA; 546 CA). No statistically significant differences were observed between races including t(11;14) (Table 3)8.
Discussion
These results suggest the presence of genetic heterogeneity between MM racial groups. Our results are consistent with the existing MM molecular literature including the observation that KRAS, NRAS, DIS3, and TP53 are commonly mutated genes in MM8. As described in many malignancies, in MM, TP53 mutations indicate a poor prognosis and shorter survival; however, the effects of other mutations are not well characterized21,22. We observed TP53 mutations more frequently in CA, which is consistent with the findings from a recent study that also observed significantly higher TP53 mutation rates among CA MM cases8. Located on chromosome 17p13.1, TP53 encodes for p53 tumor suppressor protein mediating multiple cell cycle pathways including apoptosis, cell cycle arrest, and inhibition of angiogenesis23. In MM, TP53 mutations are a rare occurrence at diagnosis; however, the incidence increases as patients are treated. It is often associated with poor prognosis and accounts for a significantly lower survival rate24. This finding suggests potentially one etiology for the worse prognosis observed in CA4. Furthermore, this may have important clinical implications in terms of targeted drug development. For example, compounds in various stages of development including MDM2 inhibitors, focus on restoring wild-type p53 activity25. A few of these agents have proceeded to first-in-human phase 1 interventional MM clinical trials26.
Importantly, in our cohort, t(11;14) was found to be significantly more frequent in the AA group (p = 0.0005). t(11;14) is a frequent translocation in MM found in about 15–20% of patients10,27,28. Recently, the Mayo Clinic group using traditional FISH methods observed a similar association with t(11;14) in their AA cohort29. They evaluated 881 patients with monoclonal gammopathies and found that the probability of having t(11;14) (or t(14;16)/t(14;20) was significantly higher in the 120 patients with highest AA ancestry (≥80%) compared with individuals with lowest levels of AA ancestry. This finding helps to confirm that our results are not random, and that NGS methods can be used to confirm traditional FISH findings. The t(11;14)(q13;q32) results in upregulation of cyclin D1, thus promoting cell cycle progression10. Most data support that the presence of t(11;14) is associated with neutral or standard prognostic risk, and that it may confer improved survival and response to treatment compared with the other commonly observed IgH translocations10,28,30,31. However, a few smaller studies suggest that patients with MM harboring t(11;14) may not have the same prognosis as patients with other standard risk features32. Interestingly, recent work suggests that AA patients with t(11;14) showed a trend toward shorter median progression-free survival (PFS) compared with AA without the presence of t(11;14); however, t(11;14) did not impact PFS in non-AA patients33. More importantly, this genetic alteration also has significant implications for drug development. More recently, it has been observed that patients with this translocation are much more likely to respond to BLC-2 inhibitors, and that this genotype is associated with increased expression of the anti-apoptotic protein BCL-2 compared with pro-apoptotic family members. For example, the BCL-2 inhibitor, venetoclax, as monotherapy is associated with a response rate of 40% in patients with t(11;14) compared with 21% in all comers34. In short, we were unable to confirm our finding in the CoMMpass cohort; however, the confirmatory finding in the Mayo AA cohort reinforces this finding, which has important implications in the AA population and precision drug development.
Deletion of chromosome 1p, (del(1p)), which is associated with a poor prognosis, in our cohort, appeared to also be more frequent (p = 0.037) along with amp(1q) in AA; however, these were not significant after adjusting for multiple testing and no differences were observed in the CoMMpass cohort8,35. Although t(11;14) is thought to be a primary event as it is observed in the early precursor state of MGUS, del(1p) is thought to be a secondary event further driving MM clonal evolution36. Therefore, this finding may be biased by the timing of when patients were diagnosed.
In addition to risk prognostication, the differences in somatic mutations among races may have significant implications in the development of targeted therapies. For example, the BRAFV600E mutation is a frequent and well-described driver mutation in melanoma and hairy cell leukemia with approximate response rates of 50–60% and 96–100%, respectively, to BRAF inhibition with tyrosine kinase inhibitors37,38,39. Interestingly, BRAF mutations occur in ~5% of MM cases with tumors that respond to tyrosine kinase inhibition40,41. Albeit a rare driver mutation in MM, it is an important druggable target. Interestingly, we observed a higher rate of BRAFV600E mutations in AA (14%) compared with CA (2%); however, this finding was not statistically significant and was not observed in the independent CoMMpass cohort.
Our findings along with other works suggest that the incidence of the various prognostic primary genetic events is not significantly different between races (e.g., t(4;14) and hyperdiploidy). Rather, the differences between races are predominantly events known to occur later in disease evolution. The molecular pathology of MM changes overtime and multiple clonal competitions occur in the cancer cell population through branching evolution36. Clonal and sub-clonal evolution occurs in the context of pressures present in the tumor micro-environment including treatment effect creating a branching nonlinear pathway of multiclonal MM development5. Based on this, one might speculate that the primary pathogenic events are similar across races, whereas the ensuing disease evolution follows slightly different trajectories, shaped by inter-racial differences in tumor–host interactions42.
This study is limited by the small number of patients. However, it expands upon the limited molecular data from AA with MM. We plan to further study and expand on these findings by examining the genetic alterations present and associated clinical outcome differences between races in patients with smoldering MM and comparison with NDMM. Our current actively enrolling study Carfilzomib, Lenalidomide, and Dexamethasone in High Risk Smoldering Multiple Myeloma will be the vehicle to aid in answering these important questions and thus far has shown impressive results at interim analysis (https://clinicaltrials.gov/ct2/show/NCT01572480)43,44. This information will add to our knowledge of the clonal evolution of MM, prognostic value of genetic data, and elucidate potential differences in smoldering myeloma compared with NDMM in terms of race.
Conclusions
The findings of this work significantly contribute to the understanding of molecular differences between races in MM, in a relative knowledge desert. These findings argue for more enrichment of AA patients in prospective MM treatment trials and characterization of molecular profiles.
References
Waxman, A. J. et al. Racial disparities in incidence and outcome in multiplemyeloma. a population-based study. Blood 116, 5501–5506 https://doi.org/10.1182/blood-2010-07-298760 (2010).
Greenberg, A. J. & Rajkumar, S. V. Elucidating disparities across racial and ethnic groups in multiple myeloma patients. Int. J. Hematol. 95, 453–454, https://doi.org/10.1007/s12185-012-1040-y (2012).
Costa, L. J. et al. Recent trends in multiple myeloma incidence and survival by age, race, and ethnicity in the United States. Blood Adv. 1, 282–287, https://doi.org/10.1182/bloodadvances.2016002493 (2017).
Landgren, O. et al. African-American multiple myeloma patients have a better survival than Caucasian patients: a population-based study including 28,636 patients. Blood 114, 1832 (2009).
Bolli, N. et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 5, 2997, https://doi.org/10.1038/ncomms3997 (2014).
Chapman, M. A. et al.Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472, https://doi.org/10.1038/nature09837 (2011).
Lohr, J. G. et al.Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 25, 91–101, https://doi.org/10.1016/j.ccr.2013.12.015 (2014).
Manojlovic, Z. et al.Comprehensive molecular profiling of 718 multiple myelomas reveals significant differences in mutation frequencies between African and European descent cases. PLoS Genet. 13, e1007087, https://doi.org/10.1371/journal.pgen.1007087 (2017).
Pawlyn, C. & Morgan, G. J. Evolutionary biology of high-risk multiple myeloma. Nat. Rev. Cancer 17, 543–556, https://doi.org/10.1038/nrc.2017.63 (2017).
Fonseca, R. et al. Myeloma and the t(11;14)(q13; q32); evidence for a biologically defined unique subset of patients. Blood 99, 3735–3741 (2002).
Bergsagel, P. L. & Kuehl, W. M. Molecular pathogenesis and a consequentclassification of multiple myeloma. J. Clin. Oncol 23, 6333–6338, https://doi.org/10.1200/jco.2005.05.021 (2005).
Smadja, N. Vr, Bastard, C., Brigaudeau, C., Leroux, D. & Fruchart, C. Hypodiploidy is a major prognostic factor in multiple myeloma. Blood 98, 2229–2238 (2001).
Kazandjian, D., Mailankody, S., Korde, N. & Landgren, O. Smoldering multiple myeloma: pathophysiologic insights, novel diagnostics, clinical risk models, and treatment strategies. Clin. Adv. Hematol. Oncol. 12, 578–587 (2014).
Robiou du Pont, S. et al. Genomics of multiple myeloma. J. Clin. Oncol. 35, 963–967, https://doi.org/10.1200/JCO.2016.70.6705 (2017).
Morgan, G. J., Walker, B. A. & Davies, F. E. The genetic architecture of multiple myeloma. Nat. Rev. Cancer 12, 335–348 (2012).
Walker, B. A. et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 132, 587–597 https://doi.org/10.1182/blood-2018-03-840132 (2018).
Kazandjian, D. Multiple myeloma epidemiology and survival: a unique malignancy. Semin. Oncol. 43, 676–681, https://doi.org/10.1053/j.seminoncol.2016.11.004 (2016).
Hill, E. M. et al. Molecular underpinnings of clinical disparity patterns in AfricanAmerican (AA) versus Caucasian American (CA) multiple myeloma (MM)patients. J. Clin. Oncol 36, 8036, https://doi.org/10.1200/JCO.2018.36.15_suppl.8036 (2018).
Malin Hultcrantz, V. Y., et al. myTYPE targeted next generation sequencing assay for detection of IgH translocations and copy number alterations in multiple myeloma https://learningcenter.ehaweb.org/eha/2018/stockholm/214976/malin.hultcrantz.mytype.targeted.next.generation.sequencing.assay.for.html (Abstract PF525 presented at European Hematology Association Annual Meeting, Stockholm. EHA Learning Center. 2018; 214976).
Lonial, S. et al.Interim analysis of the Mmrf Commpass Trial: identification of novel rearrangements potentially associated with disease initiation and progression. Blood 124, 722–722 (2014).
Alexandrov, L. B. et al.Signatures of mutational processes in human cancer. Nature 500, 415–421, https://doi.org/10.1038/nature12477 (2013).
Walker, B. A. et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J. Clin. Oncol. 33, 3911–3920, https://doi.org/10.1200/jco.2014.59.1503 (2015).
Vousden, K. H. & Lu, X. Live or let die: the cell’s response to p53. Nat. Rev. Cancer 2, 594–604, https://doi.org/10.1038/nrc864 (2002).
Chng, W. J. et al.Clinical significance of TP53 mutation in myeloma. Leukemia 21, 582–584, https://doi.org/10.1038/sj.leu.2404524 (2007).
Zhao, D., Tahaney, W. M., Mazumdar, A., Savage, M. I. & Brown, P. H. Molecularly targeted therapies for p53-mutant cancers. Cell. Mol. Life Sci. 74, 4171–4187, https://doi.org/10.1007/s00018-017-2575-0 (2017).
Tisato, V., Voltan, R., Gonelli, A., Secchiero, P. & Zauli, G. MDM2/X inhibitors under clinical evaluation: perspectives for the management of hematological malignancies and pediatric cancer. J. Hematol. Oncol. 10, 133, https://doi.org/10.1186/s13045-017-0500-5 (2017).
Sawyer, J. R.The prognostic significance of cytogenetics and molecular profiling in multiple myeloma. Cancer Genet. 204, 3–12, https://doi.org/10.1016/j.cancergencyto.2010.11.002 (2011).
Avet-Loiseau, H. et al.Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood 109, 3489–3495, https://doi.org/10.1182/blood-2006-08-040410 (2007).
Baughn, L. B. et al.Differences in genomic abnormalities among African individuals with monoclonal gammopathies using calculated ancestry. Blood Cancer J. 8, 96, https://doi.org/10.1038/s41408-018-0132-1 (2018).
Moreau, P. et al. Recurrent 14q32 translocations determine the prognosis of multiple myeloma, especially in patients receiving intensive chemotherapy. Blood 100, 1579–1583, https://doi.org/10.1182/blood-2002-03-0749 (2002).
Leiba, M. et al. Translocation t(11;14) in newly diagnosed patients with multiple myeloma: Is it always favorable?. Genes Chromosomes Cancer 55, 710–718, https://doi.org/10.1002/gcc.22372 (2016).
Lakshman, A. et al.Natural history of t(11;14) multiple myeloma. Leukemia 32, 131–138, https://doi.org/10.1038/leu.2017.204 (2018).
Gasparetto, C. J. et al.Differential effect of t(11;14) abnormality on survival and depth of response in African American (AA) and non-AA (NAA) patients (Pts) with newly diagnosed multiple myeloma (NDMM) in the Connect® MM Registry. Blood 130, 3101–3101 (2017).
Kortüm, K. M. & Einsele, H. First targeted therapy in multiple myeloma. Blood 130, 2359–2360, https://doi.org/10.1182/blood-2017-09-805341 (2017).
Ouyang, J., Gou, X., Ma, Y., Huang, Q. & Jiang, T.Prognostic value of 1p deletion for multiple myeloma: a meta-analysis. Int. J. Lab. Hematol. 36, 555–565, https://doi.org/10.1111/ijlh.12189 (2014).
Manier, S. et al.Genomic complexity of multiple myeloma and its clinical implications. Nat. Rev. Clin. Oncol. 14, 100–113, https://doi.org/10.1038/nrclinonc.2016.122 (2017).
Chapman, P. B. et al.Improved survival with Vemurafenib in melanoma with BRAF V600E mutation. New Engl. J. Med. 364, 2507–2516, https://doi.org/10.1056/NEJMoa1103782 (2011).
Hauschild, A. et al.Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet 380, 358–365, https://doi.org/10.1016/s0140-6736(12)60868-x (2012).
Tiacci, E. et al.Targeting mutant BRAF in relapsed or refractory hairy-cell leukemia. New Engl. J. Med. 373, 1733–1747, https://doi.org/10.1056/NEJMoa1506583 (2015).
Andrulis, M. et al.Targeting the BRAF V600E mutation in multiple myeloma. Cancer Discov. 3, 862–869, https://doi.org/10.1158/2159-8290.Cd-13-0014 (2013).
Raje, N. et al.Vemurafenib (VEM) in relapsed refractory multiple myeloma harboring BRAFV600 mutations (V600m): a cohort of the Histology-Independent VE-Basket Study. Blood 126, 4263–4263 (2015).
Özdemir, B. C. & Dotto, G.-P. Racial differences in cancer susceptibility andsurvival: more than the color of the skin?. Trends Cancer 3, 181–197, https://doi.org/10.1016/j.trecan.2017.02.002 (2017).
Kazandjian, D. et al. Sustained minimal residual disease negativity in newly diagnosed multiple myeloma (NDMM) patients treated with Carfilzomib (CFZ), Lenalidomide (LEN), and Dexamethasone (DEX) followed by 2 years of lenalidomide maintenance (CRd-R): updated results of a phase 2 study. Blood 128, 4527–4527 (2016).
Mailankody, S. et al. Baseline mutational patterns and sustained MRD negativityin patients with high-risk smoldering myeloma. Blood Adv 1, 1911–1918, https://doi.org/10.1182/bloodadvances.2017005934 (2017).
Acknowledgements
We thank Dr. Martin Mendoza and the Office of Minority Affairs, U.S. Food and Drug Administration and the Memorial Sloan Kettering Core Grant (P30 CA008748) for grant support in conducting this work. The views expressed in this manuscript are those of the author and do not reflect the official policy of the Department of Army/Navy/Air Force, Department of Defense, or U.S. Government.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kazandjian, D., Hill, E., Hultcrantz, M. et al. Molecular underpinnings of clinical disparity patterns in African American vs. Caucasian American multiple myeloma patients. Blood Cancer Journal 9, 15 (2019). https://doi.org/10.1038/s41408-019-0177-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41408-019-0177-9
This article is cited by
-
Understanding Myeloma-Related Information Needs and Communication Preferences Within Black American Communities: An Exploratory Study
Journal of Cancer Education (2024)
-
Addressing the disparities: the approach to the African American patient with multiple myeloma
Blood Cancer Journal (2023)
-
Black patients with multiple myeloma have better survival than white patients when treated equally: a matched cohort study
Blood Cancer Journal (2022)
-
Differences in the cytogenetic underpinnings of AL amyloidosis among African Americans and Caucasian Americans
Blood Cancer Journal (2022)
-
Isabl Platform, a digital biobank for processing multimodal patient data
BMC Bioinformatics (2020)