Abstract
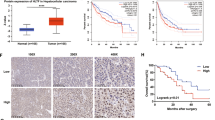
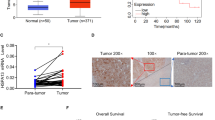
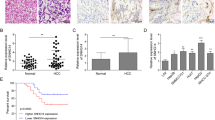
Heat shock protein family A member 8 (HSPA8) participates in the folding or degradation of misfolded proteins under stress and plays critical roles in cancer. In this study, we investigated the function of HSPA8 in the development of liver cancer. By analyzing the TCGA transcriptome dataset, we found that HSPA8 was upregulated in 134 clinical liver cancer tissue samples, and positively correlated with poor prognosis. IHC staining showed the nuclear and cytoplasmic localization of HSPA8 in liver cancer cells. Knockdown of HSPA8 resulted in a decrease in the proliferation of HepG2 and Huh-7 cells. ChIP-seq and RNA-seq analysis revealed that HSPA8 bound to the promoter of pleckstrin homology-like domain family A member 2 (PHLDA2) and regulated its expression. The transcription factor ETV4 in HepG2 cells activated PHLDA2 transcription. HSPA8 and ETV4 could interact with each other in the cells and colocalize in the nucleus. From a functional perspective, we demonstrated that HSPA8 upregulated PHDLA2 through the coactivating transcription factor ETV4 to enhance the growth of liver cancer in vitro and in vivo. From a therapeutic perspective, we identified both HSPA8 and PHDLA2 as novel targets in the treatment of HCC. In conclusion, this study demonstrates that HSPA8 serves as a coactivator of ETV4 and upregulates PHLDA2, leading to the growth of HCC, and is a potential therapeutic target in HCC treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sangro B, Melero I, Wadhawan S, Finn RS, Abou-Alfa GK, Cheng AL, et al. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol. 2020;73:1460–9.
Liu C, Lou W, Yang JC, Liu L, Armstrong CM, Lombard AP, et al. Proteostasis by STUB1/HSP70 complex controls sensitivity to androgen receptor targeted therapy in advanced prostate cancer. Nat Commun. 2018;9:4700–16.
Bonam SR, Ruff M, Muller S. Hspa8/hsc70 in immune disorders: a molecular rheostat that adjusts chaperone-mediated autophagy substrates. Cells. 2019;8:849–74.
Rai R, Kennedy AL, Isingizwe ZR, Javadian P, Benbrook DM. Similarities and differences of HSP70, HSC70, GRP78, and mortalin as cancer biomarkers and drug targets. Cells. 2021;10:2996–3014.
Dong J, Wu Z, Wang D, Pascal LE, Nelson JB, Wipf P, et al. Hsp70 binds to the androgen receptor N-terminal domain and modulates the receptor function in prostate cancer cells. Mol Cancer Ther. 2019;18:39–50.
Tanaka M, Mun S, Harada A, Ohkawa Y, Inagaki A, Sano S, et al. Hsc70 contributes to cancer cell survival by preventing rab1a degradation under stress conditions. PLoS One. 2014;9:e96785–95.
Matsuda Y, Ishiwata T, Yoshimura H, Hagio M, Arai T. Inhibition of nestin suppresses stem cell phenotype of glioblastomas through the alteration of post-translational modification of heat shock protein HSPA8/HSC71. Cancer Lett. 2015;357:602–11.
Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182:1044–61.
Albakova Z, Armeev GA, Kanevskiy LM, Kovalenko EI, Sapozhnikov AM. Hsp70 multi-functionality in cancer. Cells. 2020;9:587–612.
Mishra PB, Lobo AS, Joshi KS, Rathos MJ, Kumar GA, Padigaru M. Molecular mechanisms of anti-tumor properties of p276-00 in head and neck squamous cell carcinoma. J Transl Med. 2013;11:42–52.
Razidlo GL, Wang Y, Chen J, Krueger EW, Billadeau DD, McNiven MA. Dynamin 2 potentiates invasive migration of pancreatic tumor cells through stabilization of the Rac1 GEF Vav1. Dev Cell. 2013;24:573–85.
Yang F, Xie HY, Yang LF, Zhang L, Zhang FL, Liu HY, et al. Stabilization of MORC2 by estrogen and antiestrogens through the GPER1-PRKACA-CMA pathway contributes to estrogen-induced proliferation and endocrine resistance of breast cancer cells. Autophagy. 2020;16:1061–76.
Wang B, Lan T, Xiao H, Chen ZH, Wei C, Chen LF, et al. The expression profiles and prognostic values of HSP70s in hepatocellular carcinoma. Cancer Cell Int. 2021;21:286–302.
Desideri E, Castelli S, Dorard C, Toifl S, Grazi GL, Ciriolo MR, et al. Impaired degradation of YAP1 and IL6ST by chaperone-mediated autophagy promotes proliferation and migration of normal and hepatocellular carcinoma cells. Autophagy. 2022;19:1–11.
Khosla R, Hemati H, Rastogi A, Ramakrishna G, Sarin SK, Trehanpati N. Mir-26b-5p helps in EpCAM+cancer stem cells maintenance via HSC71/HSPA8 and augments malignant features in hcc. Liver Int. 2019;39:1692–703.
Reitman ZJ, Paolella BR, Bergthold G, Pelton K, Becker S, Jones R, et al. Mitogenic and progenitor gene programmes in single pilocytic astrocytoma cells. Nat Commun. 2019;10:3731–47.
Feng Y, Pauklin S. Revisiting 3d chromatin architecture in cancer development and progression. Nucleic Acids Res. 2020;48:10632–47.
Yuan H, Zhao L, Yuan Y, Yun H, Zheng W, Geng Y, et al. HBx represses WDR77 to enhance HBV replication by DDB1-mediated WDR77 degradation in the liver. Theranostics. 2021;11:8362–78.
Heidari Z, Chrisman IM, Nemetchek MD, Novick SJ, Blayo AL, Patton T, et al. Definition of functionally and structurally distinct repressive states in the nuclear receptor ppargamma. Nat Commun. 2019;10:5825–39.
Chittori S, Hong J, Bai Y, Subramaniam S. Structure of the primed state of the atpase domain of chromatin remodeling factor ISWI bound to the nucleosome. Nucleic Acids Res. 2019;47:9400–9.
Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–51.
Haberle V, Arnold CD, Pagani M, Rath M, Schernhuber K, Stark A. Transcriptional cofactors display specificity for distinct types of core promoters. Nature. 2019;570:122–6.
Wang H, Zhang S, Zhang Y, Jia J, Wang J, Liu X, et al. TAZ is indispensable for c-Myc-induced hepatocarcinogenesis. J Hepatol. 2022;76:123–34.
Piccinin E, Villani G, Moschetta A. Metabolic aspects in NAFLD, NASH and hepatocellular carcinoma: the role of PGC1 coactivators. Nat Rev Gastroenterol Hepatol. 2019;16:160–74.
Liu L, Lin J, He H. Identification of potential crucial genes associated with the pathogenesis and prognosis of endometrial cancer. Front Genet. 2019;10:373–87.
Idichi T, Seki N, Kurahara H, Fukuhisa H, Toda H, Shimonosono M, et al. Molecular pathogenesis of pancreatic ductal adenocarcinoma: Impact of passenger strand of pre-mir-148a on gene regulation. Cancer Sci. 2018;109:2013–26.
Ma Z, Lou S, Jiang Z. PHLDA2 regulates emt and autophagy in colorectal cancer via the PI3K/Akt signaling pathway. Aging (Albany NY). 2020;12:7985–8000.
Moon HG, Oh K, Lee J, Lee M, Kim JY, Yoo TK, et al. Prognostic and functional importance of the engraftment-associated genes in the patient-derived xenograft models of triple-negative breast cancers. Breast Cancer Res Treat. 2015;154:13–22.
Zhao GS, Gao ZR, Zhang Q, Tang XF, Lv YF, Zhang ZS, et al. Tssc3 promotes autophagy via inactivating the src-mediated PI3K/Akt/mTOR pathway to suppress tumorigenesis and metastasis in osteosarcoma, and predicts a favorable prognosis. J Exp Clin Cancer Res. 2018;37:188–204.
Tran KA, Pietrzak SJ, Zaidan NZ, Siahpirani AF, McCalla SG, Zhou AS, et al. Defining reprogramming checkpoints from single-cell analyses of induced pluripotency. Cell Rep. 2019;27:1726–41.
Martinez-Jimenez F, Muinos F, Sentis I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20:555–72.
Xu X, Wang B, Liu Y, Jing T, Xu G, Zhang L, et al. ETV4 potentiates nuclear yap retention and activities to enhance the progression of hepatocellular carcinoma. Cancer Lett. 2022;537:215640–53.
Losmanova T, Zens P, Scherz A, Schmid RA, Tschan MP, Berezowska S. Chaperone-mediated autophagy markers LAMP2a and HSPA8 in advanced non-small cell lung cancer after neoadjuvant therapy. Cells. 2021;10:2731–45.
Xiao M, Yan M, Zhang J, Xu Q, Chen W. Carboxy-terminus HSC70 interacting protein exerts a tumor inhibition function in head and neck cancer. Oncol Rep. 2017;38:1629–36.
Cancer Genome Atlas Research Network. Electronic address wbe, Cancer Genome Atlas Research N. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–41.
Wang Y, Zhao M, Zhao L, Geng Y, Li G, Chen L, et al. Hbx-induced HSPA8 stimulates HBV replication and suppresses ferroptosis to support liver cancer progression. Cancer Res. 2023;83:1048–61.
Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–67.
Zhao M, Bu Y, Feng J, Zhang H, Chen Y, Yang G, et al. Spin1 triggers abnormal lipid metabolism and enhances tumor growth in liver cancer. Cancer Lett. 2020;470:54–63.
Zheng R, Wan C, Mei S, Qin Q, Wu Q, Sun H, et al. Cistrome data browser: Expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019;47:D729–D735.
Baldavira CM, Machado-Rugolo J, Prieto TG, Bastos DR, Balancin M, Ab’Saber AM, et al. The expression patterns and prognostic significance of pleckstrin homology-like domain family a (PHLDA) in lung cancer and malignant mesothelioma. J Thorac Dis. 2021;13:689–707.
Ruart M, Chavarria L, Camprecios G, Suarez-Herrera N, Montironi C, Guixe-Muntet S, et al. Impaired endothelial autophagy promotes liver fibrosis by aggravating the oxidative stress response during acute liver injury. J Hepatol. 2019;70:458–69.
Lahiri V, Hawkins WD, Klionsky DJ. Watch what you (self-) eat: Autophagic mechanisms that modulate metabolism. Cell Metab. 2019;29:803–26.
Filali-Mouncef Y, Hunter C, Roccio F, Zagkou S, Dupont N, Primard C, et al. The menage a trois of autophagy, lipid droplets and liver disease. Autophagy. 2022;18:50–72.
Li J, Ge Z. High HSPA8 expression predicts adverse outcomes of acute myeloid leukemia. BMC Cancer. 2021;21:475–85.
Tian Y, Xu H, Farooq AA, Nie B, Chen X, Su S, et al. Maslinic acid induces autophagy by down-regulating HSPA8 in pancreatic cancer cells. Phytother Res. 2018;32:1320–31.
Kim E, Kim D, Lee JS, Yoe J, Park J, Kim CJ, et al. Capicua suppresses hepatocellular carcinoma progression by controlling the ETV4-MMP1 axis. Hepatology. 2018;67:2287–301.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 82103066) and the Natural Science Foundation of Tianjin Science and Technology Committee (19YFZCSY00020).
Author information
Authors and Affiliations
Contributions
SW, YFW, and GY performed most of the experiments; HHZ, HFY, CYH, LNZ, YHS, JS, LLS, and PL accomplished some of the in vitro and in vivo experiments; XDZ, WL, NNZ, and YS conceived and designed the project and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, S., Wang, Yf., Yang, G. et al. Heat shock protein family A member 8 serving as a co-activator of transcriptional factor ETV4 up-regulates PHLDA2 to promote the growth of liver cancer. Acta Pharmacol Sin 44, 2525–2536 (2023). https://doi.org/10.1038/s41401-023-01133-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-023-01133-3