Abstract
Nuclear factor kappa B (NF-κB) activation contributes to many vascular inflammatory diseases. The present study tested the hypothesis that microRNA-17-3p (miR-17-3p) suppresses the pro-inflammatory responses via NF-κB signaling in vascular endothelium. Human umbilical vein endothelial cells (HUVECs), transfected with or without miR-17-3p agomir/antagomir, were exposed to lipopolysaccharide (LPS), and the inflammatory responses were determined. The cellular target of miR-17-3p was examined with dual-luciferase reporter assay. Mice were treated with miR-17-3p agomir and the degree of LPS-induced inflammation was determined. In HUVECs, LPS caused upregulation of miR-17-3p. Overexpression of miR-17-3p in HUVECs inhibited NIK and IKKβ binding protein (NIBP) protein expression and suppressed LPS-induced phosphorylation of inhibitor of kappa Bα (IκBα) and NF-κB-p65. The reduced NF-κB activity was paralleled by decreased protein levels of NF-κB-target gene products including pro-inflammatory cytokine [interleukin 6], chemokines [interleukin 8 and monocyte chemoattractant protein-1] and adhesion molecules [vascular cell adhesion molecule-1, intercellular adhesion molecule-1 and E-selectin]. Immunostaining revealed that overexpression of miR-17-3p reduced monocyte adhesion to LPS-stimulated endothelial cells. Inhibition of miR-17-3p with antagomir has the opposite effect on LPS-induced inflammatory responses in HUVECs. The anti-inflammatory effect of miR-17-3p was mimicked by NIBP knockdown. In mice treated with LPS, miR-17-3p expression was significantly increased. Systemic administration of miR-17-3p for 3 days suppressed LPS-induced NF-κB activation and monocyte adhesion to endothelium in lung tissues of the mice. In conclusion, miR-17-3p inhibits LPS-induced NF-κB activation in HUVECs by targeting NIBP. The findings therefore suggest that miR-17-3p is a potential therapeutic target/agent in the management of vascular inflammatory diseases.
Similar content being viewed by others
Introduction
The endothelium, a monolayer of cells lining the interior surface of blood vessels, plays a central role in the development of vascular inflammatory diseases such as sepsis and atherosclerosis [1,2,3,4]. In response to inflammatory stimuli, such as lipopolysaccharide (LPS), tumor necrosis factor α (TNFα) and oxidized low-density lipoprotein, endothelial cells express adhesion molecules including selectins, vascular cell adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1), which mediate the tethering and rolling of monocytes on the endothelial surface [5,6,7]. Activated endothelial cells also produce chemotactic factors to facilitate the transmigration of monocytes, which differentiate into macrophages, across the vascular wall [5, 6]. The infiltrated macrophages further enhance the expression of adhesion molecules and the release of chemokines and cytokines by endothelial cells, thereby aggravating the inflammatory conditions [5,6,7,8].
Activated endothelial cells exhibit an enhanced activity of nuclear factor kappa B (NF-κB) [9, 10], which is a major transcription factor regulating inflammatory and immune responses [11, 12]. In the unstimulated state, NF-κB resides in the cytoplasm in an inactive form through binding to the inhibitors of kappa B (IκBs) [13, 14]. Upon stimulation, IκB kinase (IKK) is activated to phosphorylate the NF-κB-bound IκB. The phosphorylated IκB undergoes proteasomal degradation, and the released NF-κB dimer (mainly p65/p50) translocates to the nucleus [13, 14]. An alternative pathway for NF-κB activation involves the processing of the NF-κB subunit p100 by NF-κB inducing kinase (NIK) and the subsequent translocation of the resulted dimer RelB/p52 to the nucleus [15,16,17]. Inside the nucleus, NF-κB mediates the transcription of the genes encoding pro-inflammatory cytokines [such as interleukin (IL) 6] and cell adhesion molecules (such as E-selectin, VCAM-1 and ICAM) [18]. Therefore, inhibition of NF-κB activity in endothelial cells can be an effective approach to reduce the severity of vascular inflammatory diseases.
MicroRNA (miRNA) is a small non-coding RNA molecule with about 21–24 nucleotides (nt) and induces potent gene silencing by complementarily binding to the 3′ untranslated region (UTR) of mRNA, thus having a crucial role in many cellular and biological processes [19,20,21]. The miR-17~92 is one of the best-characterized polycistronic miRNA clusters. It encodes six individual miRNAs, miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a, and contributes to the regulation of adipocyte differentiation [22], and heart and lung development [23, 24]. Despite the general consensus that passenger miRNAs have no regulatory activity, emerging evidence demonstrated that the physiological relevance of the passenger miRNAs of the miR-17~92 cluster has been underestimated [25,26,27]. In particular, miR-17-3p (a passenger miRNA of miR-17) promotes cardiomyocyte proliferation [25], oxidative stress [26] and cell apoptosis [27]. In endothelial cells, miR-17-3p is induced following TNFα stimulation, and targets the 3′UTR of ICAM-1 mRNA to reduce the translation of the latter to protein, subsequently reducing the adhesion of leukocytes to endothelial monolayer; as such, it serves as a mechanism to limit the inflammatory responses [28]. This study aimed to examine the hypothesis that miR-17-3p is an important regulator of endothelial activation during inflammatory responses to bacterial infection and that the underlying mechanism involves modulation of a broader NF-κB signaling cascade.
Materials and methods
Cell culture
Human umbilical vein endothelial cells (HUVECs) and THP-1 monocytic cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). HUVECs were cultured in Ham’s Kaighn’s Modification F12K medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen), 1% penicillin/streptomycin (100 U·mL−1, Invitrogen), heparin (15 IU·mL−1, LEO Pharma, Denmark) and vascular endothelial growth factor (30 ng·mL−1, Sigma, St. Louis, MO, USA). THP-1 cells were cultured in Roswell Park Memorial Institute 1640 (RPMI-1640) medium (ATCC) supplemented with 10% FBS, 2-mercaptoethanol (0.05 mM, Invitrogen) and 1% penicillin/streptomycin. All cells were incubated at 37 °C in an atmosphere containing 5% CO2 −95% room air.
HUVEC transfection
HUVECs were transfected with micrON hsa-miR-17-3p agomir (17-3p, 100 nM, RIBOBIO, Guangzhou, Guangdong, China), micrOFF hsa-miR-17-3p antagomir (Anti-17-3p, 100 nM, RIBOBIO) or small interfering RNA (siRNA) against NIK and IKKβ binding protein (NIBP, 100 nM, Invitrogen) without/with co-transfection with Anti-17-3p (100 nM) using Lipofectamine 2000 (Invitrogen) for 5 h, followed by incubation with F12K medium for 24 h and treated with vehicle or with LPS (from E.coli. 026:B6; Sigma; 10 ng·mL−1, 2 h for immunofluorescence analysis; 4 h for RNA analysis; or 16 h for protein analysis and cell adhesion assay) before harvesting. The effects of transfection were validated by measuring the expression of miRNA-17-3p or the mRNA expression of NIBP through real-time polymerase chain reaction (RT-qPCR).
In vivo miRNA-17-3p overexpression and animal experiments
An equal volume of miRNA-17-3p agomir or its negative control (NC) (1 nmol dissolved in 100 μL sterile phosphate-buffered saline) and Lipofectamine 2000 (30 μL mixed with 70 μL phosphate-buffered saline) was mixed to form complexes, according to the manufacturer’s instructions. Each mouse (male, 8–10 weeks old) was administered 200 μL mixtures containing 1 nmol miRNA-17-3p agomir or its negative control once per day for consecutive 3 days by tail vein injection. LPS (from E.coli. 055:B5, Sigma; 40 mg·kg−1) or vehicle was injected intraperitoneally on the following day. After 4 h, mice were euthanized by overdose of pentobaribital sodium (100 mg·kg−1). The right lung was isolated for further measurement of mRNA and protein expression. The left lung was re-perfused with 10% neutral buffered formalin for immunofluorescent study. All experimental procedures were approved by The University of Hong Kong Committee on the Use of Live Animals for Teaching and Research and Institutional Animal Care and Use Committee at Harvard Medical School and carried out in compliance with the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (8th Edition, 2011; https://www.ncbi.nlm.nih.gov/books/NBK54050/).
Real-time polymerase chain reaction
Total RNA was isolated from HUVECs and lung tissue with TRIZOL reagent (Invitrogen). Equal amount of total RNA was reverse transcribed to first strand cDNA using the Superscript II RNase H Reverse Transcriptase kit (Invitrogen) for target gene detection. Specific Bulge-LoopTM miRNA primers (RIBOBIO) were used instead of the random primers in Superscript II RNase H Reverse Transcriptase kit for reverse transcription of miRNAs. cDNAs were quantified by RT-qPCR using SybrGreen Supermix (BioRad, Hercules, CA, USA) in ABI 7000 RT-qPCR detection system (Applied Biosystems, Foster City, CA, USA). The primers sequences used are shown in Table 1.
Western blotting
Cultured HUVECs or frozen lung tissue were homogenized in RIPA buffer [50 mM Tris-HCl, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 1% NP-40 and protease inhibitor cocktail tablets (one tablet per 50 mL solution), pH 7.8]. The protein concentration of the samples was determined with the Bradford assay (BioRad). The extracted protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membrane for detection with appropriate antibodies. Primary antibodies against human ICAM-1 (1:1000), human VCAM-1 (1:1000), GAPDH (1:1000), IκBα (1:1000), phospho-IκBα (1:1000), p65 (1:2000) and phospho-p65 (1:1000) were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-NIBP (1:250), anti-mouse VCAM-1 (1:1000) and anti-mouse E-selectin (1:1000) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-human E-selectin (1:1000) and anti-mouse ICAM-1 (1:1000) were purchased from R&D Systems (Minneapolis, MN, USA). Horseradish peroxidase-conjugated anti-mouse, anti-rabbit and anti-goat secondary antibodies (1:5000) were purchased from GE Healthcare (Boston, MA, USA) and DAKO (Glostrup, Denmark), respectively. Blots were visualized with AmershamTM ECLTM Western Blotting Detection Reagent (GE Healthcare) or Clarity ECL Western Blotting Detection Reagents (BioRad) and subsequently imaged by GBOX Chemi2 XR5 system (Syngene, Cambridge, UK). Image J software (National Institutes of Health, MD, USA) was used to analyze the optical densities of the immunoreactive bands. Protein presence was normalized to that of loading control (β-actin).
Immunofluorescence
HUVECs were seeded on the gelatin (0.2%)-coated coverslips in a 12-well plate. After different treatments, they were fixed with 4% paraformaldehyde and then incubated with 0.5% Triton X-100 for nuclear permeabilization. The lung tissue was fixed with 4% paraformaldehyde and embedded in paraffin. Sections of 5 μm thickness were then deparaffinized with xylene and rehydrated in water through a graded ethanol series, followed by antigen retrieval performed for 5 min in a microwave oven using 0.01 M citrate buffer. The cells or tissue sections were then blocked with 5% bovine serum albumin in phosphate-buffered saline with 0.1% Triton X-100, incubated sequentially with primary antibody [p65 (1:300, Cell Signaling Technology); F4/80 (1:500, BioLegend, San Diego, CA, USA); CD31 (1:500, R&D Systems)], secondary antibody [Rat IgG2α (1:500, BioLegend); Rabbit IgG (1:200, Invitrogen); Goat IgG (1:200, Invitrogen)] and ProLong® Gold Antifade Mountant with DAPI (Invitrogen). The images were acquired under Olympus BX41 microscope equipped with Olympus DP72 color digital camera (Olympus, Tokyo, Japan). Image analysis was performed with Image J software.
Interleukin 6, interleukin 8 and monocyte chemoattractant protein-1 measurements
After LPS or vehicle treatment, supernatant of HUVECs were collected for the quantification of IL-6, IL-8 and monocyte chemoattractant protein-1 (MCP-1) using enzyme-linked immunosorbent assay kits according to the manufacturer’s instructions (R&D Systems).
Monocyte adhesion assay
HUVECs were transfected with 17-3p, Anti-17-3p or their negative controls for 24 h and then treated with LPS (10 ng·mL−1) for 16 h. THP-1 cells were labeled with CellTrackerTM Green CMFAD (2.5 μM, Invitrogen) for 30 min at 37 °C. The labeled THP-1 cells were washed and added into transfected HUVECs for 4 h at 37 °C. Sequentially, the unbound THP-1 cells were removed and the adherent cells were fixed by 4% paraformaldehyde. Monocyte adhesion was observed using Olympus BX41 microscope equipped with Olympus DP72 color digital camera. Adherent THP-1 cells were quantified in three random fields of view per well using Image J software. Triplicate wells were analyzed for each experiment.
Argonaute2-miRNP immunoprecipitation
HUVECs were transfected with myc-Argonaute2 (AGO2) plasmid (2 μg·mL−1) using Lipofectamine 2000. After 2 h, HUVECs were then transfected with miR-17-3p or its negative control as described in the above section. The transfected HUVECs were lysed with CelLyticTM MT Cell Lysis reagent (Sigma). Cell lysates were incubated with protein A/G ultralin resin beads (Thermo Scientific, Pittsburgh, PA, USA) on ice for 30 min to remove non-specific binding and incubated overnight with anti-Myc-Tag antibody (Cell Signaling Technology) or Mouse IgG2α (BD Biosciences) under rotation at 4 °C, followed by incubation with protein A/G ultralin resin beads for 2 h under rotation at 4 °C. The immunoprecipitated RNA was extracted by TRIZOL reagent and processed for reverse transcription for determination of the miRNA level with real-time PCR.
Luciferase reporter assay
A pmiR-RB-REPORT™ vector including the 3′ UTR of human NIBP mRNA containing the putative miR-17-3p binding site was purchased from RIBOBIO. As a mutated vector, the mutation in the seed binding sites of 3′-UTR fragment of human NIBP (86–92) was generated from ACTGCAG to TGACGTC. For reporter assay, 293 T cells were cultured overnight, and then were transfected with 100 ng of wildtype or mutated plasmid and 50 nM of miR-17-3p agomir or negative control using Lipofectamine 3000 (Invitrogen) as transfection reagent according to the manufacturer’s instructions. Firefly and Renilla luciferase activities were measured 24 h post transfection using the Dual-Glo™ Luciferase Assay System (Promega, Madison, WI, USA).
Data and statistical analysis
All data are expressed as means ± standard deviations (SD) and n represents the number of experiments repeated with different batches or passages of HUVECs, or with different animals. Statistical analysis was performed using Prism 5.0 (GraphPad software, San Diego, CA, USA). Unpaired Student’s t-test or Mann–Whitney test was applied to analyze the comparison between two groups; one-way analysis of variance (ANOVA) followed by Bonferroni post hoc test was applied to compare the difference among the three treatment groups in the animal study. Two-way ANOVA followed by Bonferroni post hoc test was applied to determine individual differences between multiple groups of data. P values less than 0.05 were considered to indicate statistically significant differences.
Results
miR-17-3p suppresses LPS-induced pro-inflammatory cytokines and adhesion molecules in endothelial cells
LPS activates NF-κB signaling leading to the upregulation of pro-inflammatory cytokines and adhesion molecules in endothelial cells [18]. Upon LPS stimulation, the expression of miR-17-3p was significantly increased by 128% ± 40%, compared to unstimulated HUVECs (Fig. 1a), indicating that miR-17-3p upregulation is associated with the inflammatory responses of endothelial cells to LPS. To determine the role of miR-17-3p in endothelial activation, the effect of miR-17-3p on LPS-induced expression of pro-inflammatory mediators was examined in HUVECs by using gain- and loss-of-function experiments. The overexpression of miR-17-3p was achieved by transfection of 17-3p into HUVECs, which led to an approximately 500-fold increase of miR-17-3p level, as compared to NC-transfected HUVECs (Fig. 1b). LPS induced mRNA expressions of MCP-1, ICAM-1, VCAM-1 and E-selectin; such induction was significantly attenuated in HUVECs with overexpression of miR-17-3p by 36% ± 4%, 49% ± 5%, 54% ± 2%, and 53% ± 4%, respectively (Fig. 1c).
a Expression of miR-17-3p in HUVECs treated with vehicle or LPS (10 ng· mL−1 for 16 h), n = 6; b expression of miR-17-3p in HUVECs transfected with miR-17-3p agomir (17-3p) or its negative control (NC), n = 6; c mRNA expression of MCP-1, ICAM-1, VCAM-1 and E-selectin in NC- and 17-3p-transfected HUVECs stimulated with LPS (10 ng ·mL−1, 4 h), n = 4–6; d amounts of MCP-1, IL-6 and IL-8 released from NC- and 17-3p-transfected HUVECs stimulated with or without LPS (10 ng· mL−1, 16 h), n = 5–7; e protein levels of ICAM-1, VCAM-1 and β-actin in total extracts of NC- and 17-3p-transfected HUVECs stimulated with or without LPS (10 ng· mL−1, 16 h), n = 5; f amounts of MCP-1 protein released from miR-17-3p antagomir (Anti-17-3p)- or its negative control (Anti-NC)-transfected HUVECs stimulated with or without LPS (10 ng· mL−1, 16 h), n = 4; g protein levels of ICAM-1, VCAM-1 and β-actin in total extracts of Anti-NC- and Anti-17-3p-transfected HUVECs stimulated with or without LPS (10 ng· mL−1, 16 h), n = 3–4. Data are shown as means ± SD; unpaired Student’s t-test, Mann–Whitney test or two-way ANOVA followed by the Bonferroni post hoc test was performed. *P < 0.05 between vehicle and LPS, #P < 0.05 between NC and 17-3p or between Anti-NC and Anti-17-3p
To determine whether these mRNA expression changes are related to changes in protein levels, the protein levels of MCP-1 in cultured medium and of ICAM-1 and VCAM-1 in cell lysates were measured after HUVECs were treated with vehicle or LPS. The overexpression of miR-17-3p did not affect the basal release of MCP-1, but significantly reduced the LPS-stimulated MCP-1 level by 48% ± 8% (Fig. 1d). The protein levels of ICAM-1 and VCAM-1 in cell lysates were undetectable in unstimulated HUVECs, and induced by LPS treatment (Fig. 1e). The overexpression of miR-17-3p significantly suppressed the LPS-stimulated induction of ICAM-1 and VCAM-1 proteins by 45% ± 4% and 67% ± 13%, respectively (Fig. 1e). Moreover, the amounts of IL-6 and IL-8 released to the culture medium in response to LPS were significantly reduced by 99% ± 1% and 59% ± 6% in HUVECs with miR-17-3p overexpression (Fig. 1d). By contrast, in HUVECs transfected with Anti-17-3p, LPS-induced increases in MCP-1 (Fig. 1f), ICAM-1 and VCAM-1 (Fig. 1g) protein levels were further enhanced by 89% ± 27%, 47% ± 12%, and 170% ± 59%, respectively, when compared to LPS-treated HUVECs transfected with negative control of antagomir (Anti-NC). Taken together, these results indicate that miR-17-3p protects against endothelial inflammation by downregulating pro-inflammatory cytokine (IL-6), chemokines (IL-8 and MCP-1) and adhesion molecules (ICAM-1 and VCAM-1).
miR-17-3p inhibits monocyte adhesion to activated endothelial cell monolayers
During inflammatory responses, endothelial cell surface expresses adhesion molecules, such as ICAM-1, VCAM-1 and E-selectin, to promote monocyte attachment and rolling [5, 6]. To determine the functional consequence of the effects of miR-17-3p on adhesion molecule expressions, in vitro monocyte adhesion assay was performed to evaluate monocyte-endothelial cell interactions. As anticipated, the attachment of THP-1 monocytes onto HUVECs transfected with miR-17-3p agomir, antagomir or their respective negative control was minimal under basal conditions (Fig. 2). LPS stimulated the adhesion of THP-1 monocytes to HUVECs; the adhesion was significantly reduced by 47% ± 5% in HUVECs with overexpression of miR-17-3p (Fig. 2a), and increased by 80% ± 18% in cells with inhibition of miR-17-3p (Fig. 2b). The results, thus, indicate that miR-17-3p negatively affects the LPS-induced expressions of adhesion molecules thereby preventing monocyte adhesion to endothelial cell monolayers.
a Representative field of view and quantification of THP-1 monocytes adhesion to HUVECs transfected with miR-17-3p agomir (17-3p) or its negative control (NC) and stimulated with or without LPS (10 ng· mL−1, 16 h); b representative field of view and quantification of THP-1 monocytes adhesion to miR-17-3p antagomir (Anti-17-3p)- or its negative control (Anti-NC)-transfected HUVECs stimulated with or without LPS (10 ng· mL−1, 16 h); magnification×40, scale bars: 100 μm; n = 4. Data are shown as means ± SD; two-way ANOVA followed by the Bonferroni post hoc test was performed. *P < 0.05 between vehicle and LPS, #P < 0.05 between NC and 17-3p or between Anti-NC and Anti-17-3p
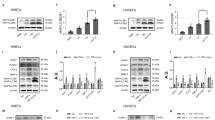
miR-17-3p reduces phosphorylation of IκBα and p65
Given the observation that in HUVECs overexpression of miR-17-3p downregulated LPS-induced expression of pro-inflammatory mediator genes, the promoter regions of which contain the NF-κB binding sites, the effects of miR-17-3p on the key events in the NF-κB signaling cascade were examined. Activation of NF-κB is initiated with the phosphorylation of its bound IκB proteins (S32/36) with the subsequent proteasomal degradation of the latter and the release of the NF-κB dimers to translocate from the cytoplasm to the nucleus [13, 14]. The phosphorylation of the NF-κB p65 subunit at serine 536 is essential for the binding of the subunit to the promoter region and hence the initiation of transcription of NF-κB-dependent genes [29]. The phosphorylation and degradation of IκBα and phosphorylation of p65 in NC- and 17-3p-transfected HUVECs, stimulated with LPS for 16 h, were examined with immunoblotting using antibodies against IκBα, phosphorylated IκBα, p65 or phosphorylated p65. The overexpression of miR-17-3p did not affect the expression of total p65, but significantly reduced the expression of phosphorylated IκBα and phosphorylated p65 in both vehicle and the LPS treatment group, as compared to NC-transfected HUVECs (Fig. 3a). In line with this, the overexpression of miR-17-3p significantly reduced the degradation of IκBα (Fig. 3a). NF-κB activity was further confirmed by p65 nuclear accumulation. A nearly 45% ± 4% reduction in p65 nuclear staining was observed in HUVECs transfected with 17-3p as compared with the cells transfected with NC (Fig. 3b). To consolidate the effect of miR-17-3p on NF-κB signaling, Anti-17-3p was transfected in HUVECs. Inhibition of miR-17-3p significantly increased the phosphorylation of IκBα (S32/36) and facilitated the degradation of IκBα in both vehicle and the LPS treatment group, as compared to Anti-NC-transfected HUVECs (Fig. 3c). Taken together, these results suggested that miR-17-3p can prevent NF-κB activation, by reducing the phosphorylation of IκBα and p65, thus preventing IκBα degradation and p65 nuclear translocation, and hence result in inhibition of NF-κB transcriptional activity.
a Protein levels of p-IκBα, IκBα, p-p65, p65 and β-actin in total extracts of HUVECs transfected with miR-17-3p agomir (17-3p) or its negative control (NC) and stimulated with or without LPS (10 ng· mL−1, 16 h), n = 4–7; b representative field of view and quantification of p65 translocation in NC- and 17-3p-transfected HUVECs stimulated with or without LPS (10 ng· mL−1, 2 h); magnification×400, scale bars: 50 μm, n = 3; c protein levels of p-IκBα, IκBα and β-actin in total extracts of miR-17-3p antagomir (Anti-17-3p)- or its negative control (Anti-NC)-transfected HUVECs stimulated with or without LPS (10 ng ·mL−1, 16 h), n = 3. Data are shown as means ± SD; two-way ANOVA followed by the Bonferroni post hoc test was performed. *P < 0.05 between vehicle and LPS, #P < 0.05 between NC and 17-3p or between Anti-NC and Anti-17-3p
miR-17-3p directly targets expression of NIK and IKKβ binding protein
To predict the potential targets of miR-17-3p, the computer program MicroCosm was used to estimate the binding energy between miRNA-17-3p and 35864 different mRNAs (Homo sapiens) in the database. ATPase family gene 3-like protein 1 (AFG3L1) and NIBP were ranked the first and second, respectively, in the order from low to high amount of the energy required for binding to miRNA-17-3p (Table 2). AFG3L1 encodes an ATP-dependent zinc metalloprotease that is target to the mitochondria in mammals [30] and there is no evidence suggesting the involvement of AFG3L1 in the regulation of inflammatory responses. However, NIBP has been reported to enhance NF-κB activity through interaction with both NIK and IKKβ [31]. Given the observation that miR-17-3p may interfere with the upstream mediators of IκBα and p65, NIBP may be the direct target of miR-17-3p for the regulation of the NF-κB signaling pathway. From the results of RNA22 program prediction, NIBP mRNA had four binding sites for miRNA-17-3p and two of them were located on the 3′UTR region (Table 3). The binding site of miRNA-17-3p was located on the positions from 75 to 92 on the 3′UTR of human NIBP mRNA (predicted by miRanda, MicroCosm and RNA22) and there were 17 pairs of Watson-Crick match (A-U pair or G-C pair) and one GU wobble (G pairing with U; Fig. 4a). For the mouse NIBP mRNA (predicted by miRanda and RNA22), the complementary sequence for miRNA-17-3p was located on the positions from 145 to 164 on the 3′UTR, with 15 pairs of Watson-Crick match and one GU wobble (Fig. 4a).
a Potential target sites for miR-17-3p binding in the 3′UTR of NIBP mRNA (human, upper; mouse, lower), as predicted by the MicroCosm, miRanda and RNA22; b mRNA level of NIBP in HUVECs co-transfected with Ago2 plasmid and miR-17-3p agomir (17-3p) or its negative control (NC), n = 3; c luciferase assay identified NIBP as a direct target gene of miR-17-3p, n = 4; d mRNA and; e protein levels of NIBP in total extracts of NC- and 17-3p-transfected HUVECs, n = 4–5; f mRNA and; g protein levels of NIBP in scramble siRNA- or NIBP siRNA (siNIBP)-transfected HUVECs, n = 4; h protein levels of p-IκBα, IκBα, p-p65, p65, ICAM-1 and β-actin in total extracts of scramble- and siNIBP-transfected HUVECs stimulated with or without LPS (10 ng· mL−1, 16 h), n = 3; i mRNA level of ICAM-1 in scramble- and siNIBP-transfected HUVECs stimulated with or without LPS (10 ng· mL−1, 4 h), n = 3; j protein levels of p-p65, p65 and GAPDH in total extracts of HUVECs transfected with Anti-17-3p without or with siNIBP and stimulated with or without LPS (10 ng· mL−1, 16 h), n = 4; data are shown as means ± SD; unpaired Student’s t-test, Mann–Whitney test, one-way or two-way ANOVA followed by the Bonferroni post hoc test was performed. *P < 0.05 between vehicle and LPS, #P < 0.05 between NC and 17-3p or between scramble and siNIBP or between Anti-NC and Anti-17-3p, $P < 0.05 between NIBP-WT + NC and NIBP-WT + miR-17-3p, &P <0.05 between NIBP-WT + miR-17-3p and NIBP-Mut+miR-17-3p, †P < 0.05 between Anti-17-3p + scramble and Anti-17-3p + siNIBP
To verify that miR-17-3p directly targets NIBP, AGO2 micro-ribonucleoprotein immunoprecipitation study was performed to examine whether NIBP mRNA is enriched in the RNA-induced silencing complex following miR-17-3p overexpression. An approximately 2.5-fold increase of NIBP was observed after AGO2 micro-ribonucleoprotein immunoprecipitation in the presence of overexpression of miR-17-3p, as compared to HUVECs transfected with NC (Fig. 4b), suggesting that NIBP is the direct target of miR-17-3p. Luciferase reporter assay further confirmed the direct interaction between miR-17-3p and NIBP, as evidenced by the significant reduction in luciferase activity for the co-transfection with wild-type NIBP plasmid and miR-17-3p when compared with the control group. However, such reduction of luciferase activity was absent, and the luciferase activity was even significantly increased by 30% ± 2% when the binding site was mutated, indicating that NIBP is a direct target gene of miR-17-3p (Fig. 4c). Furthermore, although the mRNA level of NIBP was not altered by miR-17-3p overexpression (Fig. 4d), the protein level of NIBP was significantly reduced in 17-3p-transfected HUVECs when compared with NC-transfected HUVECs (Fig. 4e); the results thus suggest that miR-17-3p targets NIBP through translation inhibition but not mRNA degradation. To further confirm whether knockdown of NIBP mimics the inhibitory effect of miR-17-3p on NF-κB signaling, siRNA was employed to reduce endogenous NIBP levels in HUVECs. A significant reduction of 57% ± 1% and 74% ± 8% in NIBP mRNA and protein level, respectively, was achieved in HUVECs transfected with NIBP siRNA as compared to mock-transfected cells (Fig. 4f, g). The knockdown of NIBP significantly reduced LPS-stimulated phosphorylation of IκBα and p65 and degradation of IκBα (Fig. 4h), and suppressed the mRNA and protein levels of NF-κB-mediated ICAM-1 in LPS-treated HUVECs (Fig. 4h, i). To strengthen the notion that reduction of NIBP accounts for the anti-inflammatory effect of miR-17-3p, the effects of LPS on NF-κB signaling were examined in HUVECs transfected with miR-17-3p antagomir without and with co-transfection with NIBP siRNA. Transfection of miR-17-3p antagomir significantly enhanced LPS-stimulated phosphorylation of p65; the enhancement was prevented by co-transfection with NIBP siRNA (Fig. 4j). Collectively, these data indicate that miR-17-3p inhibits the NF-κB signaling pathway by directly targeting NIBP expression.
miR-17-3p suppresses LPS-induced NF-κB activation and its mediated expression of adhesion molecules in vivo
Activation of endothelial cells plays a crucial role in releasing pro-inflammatory cytokines and adhesion molecules and the subsequent recruitment of neutrophils and macrophages on the endothelial surface during disease states, such as sepsis and atherosclerosis [5,6,7]. Inflammatory responses can be induced in mice with an intraperitoneal injection with LPS [32]. To evaluate the role of miR-17-3p in inflammatory responses in vivo, the level of this miRNA in the lung and the effect of overexpression of this miRNA on the recruitment of monocytes to the lungs were determined in the LPS-treated mice. In line with the in vitro findings, the expression of miR-17-3p in the lung tissue was significantly increased by 97% ± 21% after LPS treatment, and was further increased by systemic administration of 17-3p by ~20 fold as compared to NC-treated mice (Fig. 5a). In addition, LPS injection significantly increased the protein expression of NIBP, whilst overexpression of miR-17-3p significantly attenuated the NIBP level in the lung tissue (Fig. 5b). At 4 h after LPS injection, the mRNA levels of ICAM-1, VCAM-1 and E-selectin in the lung were significantly increased; such induction was significantly suppressed in the lungs of mice treated with 17-3p by 33% ± 9%, 46% ± 10% and 37% ± 17%, respectively (Fig. 5c). To determine whether these mRNA expression changes are translated to the protein level, expression of ICAM-1, VCAM-1 and E-selectin in lung tissues was quantified by Western analyses. LPS treatment significantly increased the protein levels of the adhesion molecules, and the systemic administration of 17-3p significantly reduced the LPS-induced protein expression of ICAM-1 and E-selectin by 21% ± 6% and 32% ± 7%, respectively, but not the VCAM-1 level (Fig. 5d). Analysis of lung sections taken from mice treated with 17-3p revealed a significant reduction of the numbers of F4/80-positive (monocyte/macrophage-specific marker) cells adherent to the lung endothelium (CD31 staining, endothelial cell-specific marker) in response to LPS treatment (by 46% ± 6%, Fig. 5e), thus suggesting that miR-17-3p attenuates monocyte adherence to the endothelium in vivo. Moreover, in mice challenged with LPS, there was significantly increased expression of phosphorylated IκBα and reduced expression of IκBα in the lung tissue when compared with vehicle-treated mice (Fig. 5d). Systemic administration of miR-17-3p significantly augmented the protein level of IκBα in LPS-treated mice (by 47% ± 17%, Fig. 5d). Taken together, these results demonstrate that miR-17-3p plays a critical role in LPS-induced endothelial inflammation and suppresses NF-κB-mediated expression of adhesion molecules and the subsequent monocyte adherence in vivo.
a Expression of miR-17-3p in the lung tissue of mice treated without (Vehicle) or with miR-17-3p agomir (17-3p) or its negative control (NC) and with LPS treatment (i.p. 40 mg·kg−1), n = 6–7; b protein levels of NIBP and GAPDH in total extracts of lung tissue of mice treated with NC or 17-3p with LPS treatment, n = 4; c mRNA levels of ICAM-1, VCAM-1 and E-selectin in the lung tissue of mice treated with NC or 17-3p with LPS treatment, n = 6–7; d protein levels of ICAM-1, VCAM-1, E-selectin, p-IκBα, IκBα and β-actin in total extracts of lung tissue of mice treated with NC or 17-3p with LPS treatment, n = 6–7; e representative field of view and quantification of immunofluorescent staining of macrophage (F4/80), endothelial cell (CD31) and nucleus (DAPI) in adjacent sections of lung tissue of mice treated with NC or 17-3p with LPS treatment, magnification×400, scale bars: 60 μm; n = 5. Data are shown as means ± SD; one-way ANOVA followed by the Bonferroni post hoc test was performed. *P < 0.05 between vehicle and LPS, #P < 0.05 between NC and 17-3p
Discussion
Sustained endothelial inflammation has a detrimental effect on vascular function, and contributes to the pathogenesis of both acute (such as sepsis) and chronic inflammatory diseases (such as atherosclerosis) [33, 34]. The present study demonstrated that, both in cultured human endothelial cells and in mice, miR-17-3p was upregulated by the inflammatory stimulus, the bacterial endotoxin LPS, and the upregulation was associated with reduced LPS-induced production of pro-inflammatory cytokine (IL-6), chemokines (IL-8 and MCP-1) and adhesion molecules (ICAM-1, VCAM-1 and E-selectin), and recruitment of monocytes/macrophages to endothelial cells (Fig. 6). The anti-inflammatory effect of miR-17-3p involves the inhibition of NF-κB signaling pathway activation (as demonstrated by reduced p65 nuclear translocation and phosphorylation of IκBα and p65) via directly targeting NIBP (Fig. 6). Taken in conjunction, the findings suggest that miR-17-3p is an important regulator in endothelial cells to produce self-limiting effects on inflammatory responses to pathological stimuli. Indeed, inhibition of miR-17-3p results in greater inflammatory responses (greater release of MCP-1 and expressions of ICAM-1 and VCAM-1, and enhanced phosphorylation of p65; the latter was reversed by NIBP silencing) of endothelial cells in response to LPS.
In cultured human endothelial cells, miR-17-3p was upregulated by the bacterial endotoxin LPS, and the upregulation was associated with reduced LPS-induced production of pro-inflammatory cytokine (IL-6), chemokines (IL-8 and MCP-1) and adhesion molecules (ICAM-1, VCAM-1, and E-selectin), and recruitment of monocytes/macrophages to endothelial cells. The anti-inflammatory effect of miR-17-3p involves the inhibition of NF-κB signaling pathway activation (as demonstrated by reduced p65 nucleus translocation and phosphorylation of IκBα and p65) via directly targeting NIBP
In the cytoplasm, precursor miRNA is cleaved by Dicer, an RNase-III enzyme, into a small, imperfect dsRNA that contains both the mature miRNA (“guide” or miRNA-5p) strand and its complementary (“passenger” or miRNA-3p) strand [35,36,37]. It was originally thought that the passenger miRNAs are subject to rapid degradation [36, 38, 39]. However, recent studies demonstrated that these strands could also be loaded on the Argonaute and directed the repression of the target mRNAs [40, 41]. ICAM-1 has been reported to be a direct target of miRNA-17-3p in endothelial cells [28], but this cannot explain the inhibition of NF-κB activation, the reduced release of IL-8, IL-6 or MCP-1, or the downregulation of VCAM-1 or E-selectin expression by miR-17-3p, since these effects are upstream or independent of ICAM-1 expression. Therefore, online algorithms (miRanda, MicroCosm and RNA22) were used to identify other potential mRNA target(s) that can account for the anti-inflammatory effects of miR-17-3p in LPS-stimulated endothelial cells.
Based on the bioinformatics analysis, NIBP was predicted as a direct target gene of miR-17-3p. NIBP is highly conserved across evolution (protein sequence identity between human and mouse, 92%; chicken, 87%; zebrafish, 85%; pufferfish, 75%; sea urchin, 42%; bee, 35%) [31, 42]. Through interacting with NIK and IKKβ, the upstream regulating proteins of NF-κB signaling, NIBP enhances cytokine-induced NF-κB activation in different cell lines, such as rat pheochromocytoma PC12 cells [31], human embryonic kidney HEK 293 cells [43] and human colon tumor HCT116 cells [44]. In the present study, miR-17-3p inhibited phosphorylation of p65 and IκBα, and prevented p65 translocation and IκBα degradation, thus indicating that it interferes with the activation of NF-κB pathway and is in line with NIBP being the direct target of miR-17-3p. Moreover, the sequence complementary analysis between the seed sequences of miR-17-3p and the 3′UTR of the human and mouse NIBP demonstrates, respectively, a perfect Watson-Crick match between nt 2 and 8 of the miRNA seed region (the 7mer-m8 principle) [45] and the same perfect Watson-Crick match together with an adenine-base opposite the position one at the 5′ end of miRNA (the 8mer principle) [46]; these seed sequence matching principles are considered to provide miRNA target prediction with the greatest accuracy since they have been validated with measurements of the predicted mRNA target and protein levels [46,47,48]. Further evidence supporting NIBP being the target of miR-17-3p in endothelial cells comes from the findings that (1) miR-17-3p overexpression suppressed NIBP expression at the post-transcriptional (protein) level and increased NIBP mRNA in the RNA-induced silencing complex; (2) reduced luciferase activity for the co-transfection with wildtype NIBP plasmid and miR-17-3p agomir, but had no effect when the binding site was mutated; and (3) knockdown of NIBP mRNA reproduced the anti-inflammatory effects of miR-17-3p overexpression in endothelial cells, such as reduced LPS-stimulated phosphorylation of p65 and suppressed mRNA and protein levels of ICAM-1. Taken together, these findings collectively suggested that miR-17-3p directly targets NIBP, and therefore suppresses LPS-induced activation of NF-κB signaling pathway in endothelial cells.
In addition to miR-17-3p, many miRNA networks act on NF-κB signaling pathway to inhibit inflammatory responses in endothelial cells [15, 49, 50]. For example, miR-155, which is upregulated by TNFα, directly targets p65 to suppress the NF-κB signaling pathway and reduces the adhesion of monocytes to the endothelium [49], whereas miR-181b, which is downregulated by TNFα, targets importin-α3, a protein critical for NF-κB nuclear translocation, to suppress the expression of NF-κB-regulated pro-inflammatory cytokines and adhesion molecules in vitro and in vivo [15, 51, 52]. In endothelial cells stimulated with IL-1β, the activation of the signaling pathways of NF-κB, as well as other pro-inflammatory transcriptional factors AP-1 and MAPK/EGR, is inhibited by miR-146a and miR-146b through targeting the IL-1β signaling pathway adapter proteins (such as TRAF6 and IRAK1/2) [50]. The findings confirm that a miRNA network exists to regulate the NF-κB signaling pathway at multiple levels confirms the importance of limiting the activation of this nodal transcription factor family for the maintenance of vascular homeostasis during inflammatory insults. NIBP, the target of miR-17-3p, interacts with both NIK [31] (the kinase responsible for initiating the alternative pathway for NF-κB activation involving the nuclear translocation of the NF-κB dimer RelB/p50) [15,16,17] and IKKβ [31] (the kinase responsible for initiating the classical pathway for NF-κB activation involving the nuclear translocation of mainly the NF-κB dimer p65/p52) [13, 14, 29]. The data therefore suggest that miR-17-3p can serve as an effective therapeutic agent to prevent exacerbated inflammation and the resultant tissue injury in pathological conditions involving the activation of NF-κB.
During the course of our study, Yan et al. [53] demonstrated that miR-17-3p significantly activates NF-κB pathways in human keratinocyte cell line (HaCaT), in contrast to the present finding that miR-17-3p suppresses LPS-induced activation of NF-κB pathways in HUVECs. The seemingly contradictory results may be attributed to the discrepancies between the experimental design of the two studies: (1) Different cell types were used (keratinocytes in their versus endothelial cells in the present work), and the regulatory role of miR-17-3p in NF-κB signaling pathway may be different in different cell types. Indeed, a cell-specific effect on NF-κB signaling is reported for miR-181b, which suppresses NF-κB activity in endothelial cells but not in other cell types, such as peripheral blood mononuclear cells, bone marrow-derived macrophages and peritoneal macrophages, and not in the liver [10, 51, 52]. The cell-specific effects of microRNA are likely related to the differences in the relative protein expressions in different cells; (2) Cells were exposed to different treatments. In the study of Yan et al. [53], the effect of transfecting miR-17-3p mimic to HaCaT cells, and hence the overexpression of miR-17-3p (the degree of upregulation was not indicated), on NF-κB activity under basal condition was examined. By contrast, in the present study, the effect of hsa-miR-17-3p agomir (chemically-modified double-stranded microRNA) transfection, leading to an approximately 500-fold increase of miR-17-3p level, in HUVECs on the LPS-induced activation of NF-κB was determined.
Besides regulating NF-κB activity and hence inflammatory responses, Shi et al. [25] showed that miR-17-3p controls cardiomyocyte proliferation and hypertrophy in vitro by directly targeting metallopeptidase inhibitor 3 (TIMP3) and acting upstream of the PTEN-AKT pathway, respectively. Furthermore, miR-17-3p contributes to exercise-induced cardiac growth and protects against adverse remodeling after myocardial ischemia/reperfusion injury (I/RI) [25]. However, whether miR-17-3p impacts on myocardial I/RI via TIMP3 or PTEN-AKT pathway in vivo remains unknown. Of note, during cardiac I/RI, inflammatory cells, including neutrophils and macrophages, infiltrate to the infarcted myocardium and produce an array of pro-inflammatory cytokines and chemokines via activation of NF-κB, which further exacerbate cardiomyocytes death and myocardial I/RI [54]. NF-κB activation has also been demonstrated in various models of experimental myocardial ischemia and reperfusion [55]. Moreover, miR-17-3p appears to be implicated in the inhibition by dexmedetomidine of NF-κB activation in H9C2 rat cardiomyocytes during hypoxia/reoxygenation treatment [56]. Thus, it is likely that miR-17-3p produces the beneficial effect against myocardial I/RI, at least in part, through inhibition of the NF-κB activation.
In the present study, the level of miR-17-3p was upregulated upon LPS stimulation. LPS is a major component of the outer membrane of Gram-negative bacteria [57]. It activates Toll-like receptor 4 (TLR4) in the cell membrane to initiate inflammatory responses via two major intracellular signaling pathways: one depending on the recruitment of the adapter molecule myeloid differentiation factor (MyD) and the other on that of the adapter molecule Toll/interleukin-1 receptor domain-containing adapter inducing interferon-beta (TRIF) [58]. Both MyD and TRIF recruitments to TLR4 result in the formation of a signaling complex involving different kinases and ubiquitin ligases, and subsequently lead to the upregulation of transcription of genes encoding the inflammatory mediators [58]. It is therefore logical to propose that miR-17-3p level is upregulated by LPS through the MyD or TRIF signaling complex, which modulates the activity of the key enzymes involved in the biogenesis of miR-17-3p (related to miRNA transcription, nuclear processing and maturation). The present study does not permit further speculation of the mechanism by which LPS regulates miR-17-3p expression. It appears that LPS is not the only stimulator for miR-17-3p upregulation in endothelial cells, since TNFα also increased the level of miR-17-3p in HUVEC [28].
In conclusion, the present study demonstrates that the inflammatory stimulus bacterial endotoxin can induce the expression of miR-17-3p, and that miR-17-3p has a therapeutic effect on acute inflammatory diseases, as systemic delivery of miR-17-3p in mice attenuated the inflammatory responses to a septic dose of LPS. Through binding to the 3′UTR of NIBP mRNA, miR-17-3p inhibited the activation of NF-κB signaling pathway, resulting in reduced expression of pro-inflammatory cytokines, chemokines, and cell adhesion molecules and reduced recruitment of leukocytes to the vascular endothelium. Since NF-κB activation is a major mechanism underlying chronic vascular inflammation, which contributes to the development of cardiovascular complications, including atherosclerosis and diabetes, miR-17-3p also has a therapeutic potential in the management of chronic inflammatory diseases.
References
Schöffel U, Kopp KH, Männer H, Vogel F, Mittermayer C. Human endothelial cell proliferation inhibiting activity in the sera of patients suffering from ‘shock’ or ‘sepsis’. Eur J Clin Invest. 1982;12:165–71.
McKenna TM, Martin FM, Chernow B, Briglia FA. Vascular endothelium contributes to decreased aortic contractility in experimental sepsis. Circ Shock. 1986;19:267–73.
Constantinides P. Importance of the endothelium and blood platelets in the pathogenesis of atherosclerosis. Triangle. 1976;15:53–61.
Landmesser U, Hornig B, Drexler H. Endothelial function: a critical determinant in atherosclerosis? Circulation. 2004;109:II27–33.
Lusis AJ. Atherosclerosis. Nature. 2000;407:233–41.
Libby P. Inflammation in atherosclerosis. Arterioscler Thromb Vasc Biol. 2012;32:2045–51.
Ince C, Mayeux PR, Nguyen T, Gomez H, Kellum JA, Ospina-Tascón GA, et al. The endothelium in sepsis. Shock. 2016;45:259–70.
Libby P. Inflammatory mechanisms: the molecular basis of inflammation and disease. Nutr Rev. 2007;65:S140–46.
Gareus R, Kotsaki E, Xanthoulea S, van der Made I, Gijbels MJ, Kardakaris R, et al. Endothelial cell-specific NF-kappaB inhibition protects mice from atherosclerosis. Cell Metab. 2008;8:372–83.
Sun X, Icli B, Wara AK, Belkin N, He S, Kobzik L, et al. MicroRNA-181b regulates NF-κB-mediated vascular inflammation. J Clin Invest. 2012;122:1973–90.
Xanthoulea S, Curfs DM, Hofker MH, de Winther MP. Nuclear factor kappa B signaling in macrophage function and atherogenesis. Curr Opin Lipido. 2005;16:536–42.
Cai Y, Sukhova GK, Wong HK, Xu A, Tergaonkar V, Vanhoutte PM, et al. Rap1 induces cytokine production in pro-inflammatory macrophages through NFκB signaling and is highly expressed in human atherosclerotic lesions. Cell Cycle. 2015;14:3580–92.
Ghosh S, May MJ, Kopp EB. NF-κB and rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–60.
Israël A. The IKK complex, a central regulator of NF-κB activation. Cold Spring Harb Perspect Biol. 2010;2:a000158.
Xiao G, Harhaj EW, Sun SC. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol Cell. 2001;7:401–9.
Senftleben U, Cao Y, Xiao G, Greten FR, Krähn G, Bonizzi G, et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293:1495–9.
Ramakrishnan P, Wang W, Wallach D. Receptor-specific signaling for both the alternative and the canonical NF-kappaB activation pathways by NF-kappaB-inducing kinase. Immunity. 2004;21:477–89.
Sawa Y, Ueki T, Hata M, Iwasawa K, Tsuruga E, Kojima H, et al. LPS-induced IL-6, IL-8, VCAM-1, and ICAM-1 expression in human lymphatic endothelium. J Histochem Cytochem. 2008;56:97–109.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71.
Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015;4:e252.
Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci USA. 2008;105:2889–94.
Danielson LS, Park DS, Rotllan N, Chamorro-Jorganes A, Guijarro MV, Fernandez-Hernando C, et al. Cardiovascular dysregulation of miR-17-92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J. 2013;27:1460–67.
Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–53.
Shi J, Bei Y, Kong X, Liu X, Lei Z, Xu T, et al. miR-17-3p contributes to exercise-induced cardiac growth and protects against myocardial ischemia-reperfusion injury. Theranostics. 2017;7:664–76.
Tian B, Maidana DE, Dib B, Miller JB, Bouzika P, Miller JW, et al. miR-17-3p exacerbates oxidative damage in human retinal pigment epithelial cells. PLoS ONE. 2016;11:e0160887.
Lu D, Tang L, Zhuang Y, Zhao P. miR-17-3P regulates the proliferation and survival of colon cancer cells by targeting Par4. Mol Med Rep. 2018;17:618–23.
Suárez Y, Wang C, Manes TD, Pober JS. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J Immunol. 2010;184:21–5.
Yang F, Tang E, Guan K, Wang CY. IKKβ plays an essential role in the phosphorylation of RelA/p65 on serine 536 induced by lipopolysaccharide. J Immunol. 2003;170:5630–5.
Kremmidiotis G, Gardner AE, Settasatian C, Savoia A, Sutherland GR, Callen DF. Molecular and functional analyses of the human and mouse genes encoding AFG3L1, a mitochondrial metalloprotease homologous to the human spastic paraplegia protein. Genomics. 2001;76:58–65.
Hu WH, Pendergast JS, Mo XM, Brambilla R, Bracchi-Ricard V, Li F, et al. NIBP, a novel NIK and IKK(beta)-binding protein that enhances NF-(kappa)B activation. J Biol Chem. 2005;280:29233–41.
Copeland S, Warren HS, Lowry SF, Calvano SE, Remick D. Inflammation and the host response to injury investigators. Acute inflammatory response to endotoxin in mice and humans. Clin Diagn Lab Immunol. 2005;12:60–7.
Aird WC. The role of the endothelium in severe sepsis and multiple organ dysfunction syndrome. Blood. 2003;101:3765–77.
Hansson GK, Libby P. The immune response in atherosclerosis: a double-edged sword. Nat Rev Immunol. 2006;6:508–19.
Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55.
Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68.
Rawal S, Manning P, Katare R. Cardiovascular microRNAs: as modulators and diagnostic biomarkers of diabetic heart disease. Cardiovasc Diabetol. 2014;13:44.
Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16.
Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208.
Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–63.
Winter J, Diederichs S. Argonaute-3 activates the let-7a passenger strand microRNA. RNA Biol. 2013;10:1631–43.
Mir A, Kaufman L, Noor A, Motazacker MM, Jamil T, Azam M, et al. Identification of mutations in TRAPPC9, which encodes the NIK- and IKK-beta-binding protein, in nonsyndromic autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:909–15.
Zhang Y, Bitner D, Pontes Filho AA, Li F, Liu S, Wang H, et al. Expression and function of NIK- and IKK2-binding protein (NIBP) in mouse enteric nervous system. Neurogastroenterol Motil. 2014;26:77–97.
Zhang Y, Liu S, Wang H, Yang W, Li F, Yang F, et al. Elevated NIBP/TRAPPC9 mediates tumorigenesis of cancer cells through NFκB signaling. Oncotarget. 2015;6:6160–78.
Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85.
Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105.
Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–910.
Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63.
Wu XY, Fan WD, Fang R, Wu GF. Regulation of microRNA-155 in endothelial inflammation by targeting nuclear factor (NF)-κB P65. J Cell Biochem. 2014;115:1928–36.
Cheng HS, Sivachandran N, Lau A, Boudreau E, Zhao JL, Baltimore D, et al. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol Med. 2013;5:1017–34.
Sun X, He S, Wara AKM, Icli B, Shvartz E, Tesmenitsky Y, et al. Systemic delivery of microRNA-181b inhibits nuclear factor-κB activation, vascular inflammation, and atherosclerosis in apolipoprotein E-deficient mice. Circ Res. 2014;114:32–40.
Sun X, Sit A, Feinberg MW. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc Med. 2014;24:105–12.
Yan H, Song K, Zhang G. MicroRNA-17-3p promotes keratinocyte cells growth and metastasis via targeting MYOT and regulating Notch1/NF-κB pathways. Pharmazie. 2017;72:543–9.
Frangogiannis NG. Targeting the inflammatory response in healing myocardial infarcts. Curr Med Chem. 2006;13:1877–93.
Kim JW, Jin YC, Kim YM, Rhie S, Kim HJ, Seo HG, et al. Daidzein administration in vivo reduces myocardial injury in a rat ischemia/reperfusion model by inhibiting NF-kappaB activation. Life Sci. 2009;84:227–34.
Yuan T, Yang Z, Xian S, Chen Y, Wang L, Chen W, et al. Dexmedetomidine-mediated regulation of miR-17-3p in H9C2 cells after hypoxia/reoxygenation injury. Exp Ther Med. 2020;20:917–25.
Bohl TE, Aihara H. Current Progress in the structural and biochemical characterization of proteins involved in the assembly of lipopolysaccharide. Int J Microbiol. 2018;2018:5319146.
Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–51.
Acknowledgements
This work was supported by the Health and Medical Research Fund (16151212) of the Food and Health Bureau of the Government of the Hong Kong Special Administrative Region (to SWSL), a Seed Fund for Basic Research of the University of Hong Kong (to SWSL), and the National Institutes of Health (HL115141, HL117994, HL134849, and GM115605 to MWF), the Arthur K. Watson Charitable Trust (to MWF), and the Dr. Ralph & Marian Falk Medical Research Trust (to MWF).
Author information
Authors and Affiliations
Contributions
YC, YZ, HC, and XHS conceived and designed the study, performed the experiments, analyzed the data and wrote the manuscript. PZ, LZ, MYL, and FZ performed some experiments and analyzed the data. ZYX participated in the experiment design and the interpretation of results. RYKM, MWF, and SWSL designed the experiments, analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Cai, Y., Zhang, Y., Chen, H. et al. MicroRNA-17-3p suppresses NF-κB-mediated endothelial inflammation by targeting NIK and IKKβ binding protein. Acta Pharmacol Sin 42, 2046–2057 (2021). https://doi.org/10.1038/s41401-021-00611-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-021-00611-w