Abstract
A series of 6-substituted carbazole-based retinoic acid-related orphan receptor gamma-t (RORγt) modulators were discovered through 6-position modification guided by insights from the crystallographic profiles of the “short” inverse agonist 6. With the increase in the size of the 6-position substituents, the “short” inverse agonist 6 first reversed its function to agonists and then to “long” inverse agonists. The cocrystal structures of RORγt complexed with the representative “short” inverse agonist 6 (PDB: 6LOB), the agonist 7d (PDB: 6LOA) and the “long” inverse agonist 7h (PDB: 6LO9) were revealed by X-ray analysis. However, minor differences were found in the binding modes of “short” inverse agonist 6 and “long” inverse agonist 7h. To further reveal the molecular mechanisms of different RORγt inverse agonists, we performed molecular dynamics simulations and found that “short” or “long” inverse agonists led to different behaviors of helixes H11, H11’, and H12 of RORγt. The “short” inverse agonist 6 destabilizes H11’ and dislocates H12, while the “long” inverse agonist 7h separates H11 and unwinds H12. The results indicate that the two types of inverse agonists may behave differently in downstream signaling, which may help identify novel inverse agonists with different regulatory mechanisms.
Similar content being viewed by others
Introduction
Retinoic acid receptor-related orphan receptor gamma-t (RORγt) belongs to the nuclear receptor (NR) transcription factor superfamily. It plays a key role in driving the differentiation of naive CD4+ T cells into T helper 17 (Th17) cells [1]. RORγt contains a variable N-terminal domain (A/B or AF1), a conserved DNA-binding domain (DBD), a flexible hinge region, and a C-terminal ligand-binding domain (LBD) with a ligand-dependent activation function 2 (AF2) region that recruits coactivators by interacting with helix 12 (H12). Small molecules binding to the LBD can stabilize or destabilize the activation state of AF2 to modulate the function of RORγt, thus acting as agonists or inverse agonists [2,3,4]. For years, RORγt has attracted significant attention as a drug target for its special role in the Th17/IL-17 pathway. RORγt inverse agonists such as JTE-451, ESR-114, AUR-101, and VTP-43742 for autoimmune diseases and RORγt agonists such as LYC-55716 for cancer immunotherapy are now in Phase II clinical trials, which indicates that RORγt modulators are promising in the treatment of various autoimmune diseases and cancer [5,6,7].
Some research groups accidentally discovered RORγt agonists through the study of inverse agonists with minor structural modifications [8,9,10]. Cocrystal structures of each pair of inverse agonists and agonists revealed a stable hydrophobic network (the so-called “HYF lock”) among His479 (H11), Tyr502 (H12), and Phe506 (H12) in the active state of the RORγt LBD. Recently, Takeda designed RORγt agonist 2 through a structure-based functional switching approach from inverse agonist 1 (Fig. 1a) [11]. By molecular dynamics (MD) simulation, the researchers found that the conformational change of Trp317 and the hydrogen bond interaction of His479 and Tyr502 were important in the functional switch between inverse agonists and agonists.
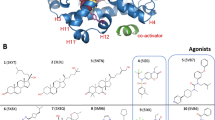
In our recent study, the functional switch phenomenon was observed between agonists and two types of inverse agonists (Fig. 1b), namely, “short” and “long” inverse agonists, which were named based on the size of the inverse agonists relative to the corresponding agonist [12]. Through removal from or addition to a minor structural moiety, RORγt agonist 4 could be converted into “short” inverse agonist 3 and “long” inverse agonist 5. Notably, the two types of RORγt inverse agonists behaved differently in cofactor peptide recruitment. The “short” inverse agonist 3 recruited corepressor peptides and dispelled coactivator peptides, while the “long” inverse agonist 5 dispelled both. However, the detailed mode of action (MOA) at the molecular level has not been studied in depth, and the underlying mechanism of “short” and “long” inverse agonists remains unclear. We believe that the difference between the two types of RORγt inverse agonists deserves a systematic study, as it not only demonstrates the regulatory mechanism of RORγt but also may guide the discovery of novel modulators with differentiated drug action and safety profiles.
Previously, we reported a series of carbazole-containing compounds, such as compound 6 (Fig. 2), as RORγt inverse agonists [13]. Herein, based on insights from the freshly resolved cocrystal structure, we classified compound 6 as a “short” inverse agonist and identified the 6-position of the carbazole moiety as the site for structural modification to achieve the functional switch from the “short” inverse agonist 6 to agonists and “long” inverse agonists. In this paper, we describe our insights into stabilizing or destabilizing the AF2 domain from cocrystal structures resolved and report the design, synthesis, and biological evaluation of 6-substituted carbazole-based RORγt modulators with substituents (R groups) gradually increasing in size to obtain RORγt “short” inverse agonists, agonists or “long” inverse agonists (7a-7j, Fig. 2). The molecular mechanisms of the RORγt agonists and two types of inverse agonists were then revealed by MD simulation studies. Different behaviors of helixes H11, H11’ and H12 of the protein with “short” or “long” inverse agonists were further elucidated.
Materials and methods
Chemical synthesis
All reagents used were commercially available and were used as received unless otherwise indicated. The microwave reaction was conducted with a Biotage Initiator™ microwave synthesizer (Biotage, Uppsala, Sweden). NMR data were recorded with a Bruker 400 MHz NMR system. Chemical shifts (δ) are expressed in parts per million (ppm) relative to tetramethylsilane (TMS) as an internal standard. Mass spectra were measured on an Agilent 1100 series LC/MSD 1947d spectrometer (Agilent Technologies, Inc., Santa Clara, CA, USA). High-resolution mass spectra (HRMS) were obtained on an AB SCIEX TripleTOF 5600+ mass spectrometer (AB SCIEX, LLC., Redwood City, CA, USA). The contents of compounds for biological evaluation were examined with an Agilent 1260 Infinity LC system (Agilent Technologies, Inc.) with methanol/water (40:60 to 95:5) as the eluent. Unless specified, the purity of the target compounds was >95%, which was considered to be pure enough for biological assays. The synthetic procedures, 1H-NMR, 13C-NMR, and HRMS are detailed in the Supporting Information section.
Compounds 6 and 7a were prepared according to the procedures described in our previous work [13]. The synthesis of the carbazole-based compounds 7b-7j is depicted in Scheme 1. Starting from commercially available 4-halogeno-nitroarenes (8 and 17) or 4-nitrophenol (15), the desired ether intermediates (9, 16, and 18) were generated through nucleophilic substitution reactions. Then, the nitro-containing intermediates were reduced to yield the corresponding anilines (10 and 19). Carbazole-based intermediates (12 and 20) were obtained via Buchwald-Hartwig cross coupling reactions, followed by palladium-catalyzed cyclization reactions. The intermediates were then alkylated, and ester hydrolysis followed by amide formation with (4-(ethylsulfonyl)phenyl)methanamine (23) resulted in the target compounds 7b-7j. The synthesized compounds were then evaluated in the RORγt dual FRET assay and Gal4 cell-based reporter gene assay.
a Reagents and conditions: (a) NaH, DMF and 1-(4-hydroxypiperidin-1-yl)ethan-1-one (21)/1-(4-hydroxypiperidin-1-yl)-2,2-dimethylpropan-1-one (22)/alkyl alcohols at 0 °C, followed by 8 at 60 °C for 3 h, yield: 49.8%–65.2%. b SnCl2·2H2O, MeOH at rt for 12 h, yield: 78.9%–92.1%. c rac-BINAP, Pd(OAc)2, K2CO3, methyl 4-bromobenzoate, and toluene under reflux for 2 h, yield: 27.3%–63.0%. d Tributylphosphonium tetrafluoroborate, Pd(OAc)2, K2CO3, and DMA under reflux for 5 h, yield: 46.8%–52.7%. e NaH and DMF at 0 °C followed by bromoethane at rt for 20 min, yield: 62.6%–90.1%. f KOH, H2O, and MeOH at 80 °C for 1 h, yield: 55.6%–100.0%. g DIPEA, HATU, (4-(ethylsulfonyl)phenyl)methanamine (23) and DCM at rt for 2 h, yield: 45.8%–88.9%. h Bromoalkanes, K2CO3 and DMF at 70 °C for 5 h, yield: 83.4%–96.2%. i Potassium tert-butoxide and THF at 60 °C for 6 h, yield: 38.6%. j Zn, HCOONH4, MeOH, and H2O at 80 °C, 2 h, yield: 61.6%–66.9%. k Pd(OAc)2 and HOAc at 130 °C, 1 h, yield: 29.8%–44.7%.
Protein expression and purification
Human nuclear receptor RORγt (residues 260–507)-GGG-SRC1 (SRC1 sequence: EKHKILHRLLQDS) was amplified by polymerase chain reaction (PCR) and then analyzed by electrophoresis on a 1% agarose gel. RORγt LBD was cloned into the His-Flag-SUMO tag pRSF DUET1 vector. The proteins were expressed in Escherichia coli strain BL21(DE3). The cells were grown in LB (Luria − Bertani broth) at 37 °C. When the OD600 nm reached 0.6, the temperature was shifted to 16 °C, and the cells were induced with 0.4 mM IPTG (isopropyl β-D-1-thiogalactopyranoside) for 16 h. The cells were collected by supercentrifugation at 4000 rpm for 15 min. RORγt protein was first purified by nickel affinity chromatography (His Trap HP, GE Healthcare, General Electric Company, NJ, USA) and then further purified by gel-filtration chromatography on a Superdex 75 10/300 column (GE Healthcare). Finally, the purified RORγt protein was concentrated in a 10 k enrichment tube (Millipore, Merck KGaA, Darmstadt, Germany) and flash frozen at −80 °C. The RORγt LBD used in the binding assay was stored at −80 °C in buffer containing 25 mM HEPES (pH = 7.4), 200 mM NaCl, and 5% glycerol. The RORγt-GGG-SRC1 protein used for crystallization was stored at −80 °C in buffer containing 25 mM HEPES (pH 7.0), 150 mM NaCl, and 2 mM dithiothreitol.
RORγt dual FRET assay
The assay was performed in an assay buffer consisting of 50 mM NaF, 50 mM 3-(N-morpholino)propanesulfonic acid (pH = 7.4), 0.05 mM 3-[(3-cholamidopropyl) dimethylammonio]propanesulfonate, 0.1 mg/mL bovine serum albumin, and 10 mM dithiothreitol in 384-well plates. The total volume was 25 μL/well. The europium-labeled SRC1 solution was prepared by adding an appropriate amount of biotinylated SRC and europium-labeled streptavidin into assay buffer, with final concentrations of 20 and 10 nM, respectively. The allophycocyanin (APC)-labeled-LBD solution was prepared by adding an appropriate amount of biotinylated RORγt-LBD and APC-labeled streptavidin at final concentrations of 20 and 10 nM, respectively. After 15 min of incubation at room temperature, a 20-fold excess of biotin was added and incubated for 10 min at room temperature to block the remaining free streptavidin. Equal volumes of europium-labeled SRC and APC-labeled RORγt-LBD were dispensed into 384-well assay plates at 25 μL volume/well. The 384-well assay plates had 100 nL of test compound in DMSO predispensed into each well. The plates were incubated for 1 h at room temperature and then read on Envision in LANCE mode configured for europeum-APC labels. Detailed data are presented in Supplementary Table S1.
RORγt GAL4 cell-based reporter gene assay
The hRORγt LBD coding sequence was inserted into a pBIND expression vector (Promega, Fitchburg, WI, USA) to express ROR-GAL4 binding domain chimeric receptors. This expression vector and a reporter vector (pGL4.35, which carries a stably integrated GAL4 promoter-driven luciferase reporter gene [luc2P/9XGAL4 UAS/Hygro]) were cotransfected into HEK293T host cells. Upon agonist binding to the corresponding ROR-GAL4 chimeric receptor, the chimeric receptor binds to the GAL4 binding sites and stimulates the reporter gene. In the presence of an inverse agonist, the agonist will bind competitively to the nuclear receptor and activate reporter gene transcription. HEK293T cells were cultured in a culture medium composed of DMEM containing 5% charcoal-treated FBS at 37 °C under a 5% CO2 atmosphere, as recommended by the ATCC. Before the assay, the cells were washed with PBS to remove phenol red and suspended in phenol red-free medium (phenol red-free DMEM containing 5% charcoal-treated FBS and penicillin-streptomycin (10,000 U/mL)) to a proper concentration. HEK293T cells (6 × 106) were seeded into a 100 mm dish and incubated for 16 h. To a reagent mixture of TransIT-293 transfection reagent (Mirus Bio, WI, USA) and Opti-MEM (Invitrogen, CA, USA), plasmid DNA (used as 0.5 mg/mL stocks) containing 5 μg RORγt plasmid and 5 μg pGL4.35 luciferase plasmid was added. The mixture was added to the cells in a 100 mm dish and incubated for 5–6 h. Test compounds were serially diluted in DMSO to 300 nM. Compounds (25 nL) were transferred to a 384-well plate (white opaque) using Echo 550. Then, the cells were seeded at 15,000 cells/well into a 384-well plate using phenol red-free DMEM containing 5% charcoal-treated FBS and 0.25 μM ursolic acid. Cells were incubated for 16–20 h at 37 °C (white opaque) using Echo 550. Then, 25 μL of SteadyGlo™ Luciferase Assay Reagent (Promega) was added to each well of the 384-well plate. The plate (avoiding light) was shaken for 5 min on a plate shaker (4625-1CECN/THZ Q, Thermo Fisher Scientific, MA, USA). The luminescence value was recorded on an Envision 2105 plate reader (PerkinElmer, MA, USA). Activation/inhibition values of the compounds at 300 nM were determined by the average value of duplicate tests.
Microscale thermophoresis (MST) assay
The microscale thermophoresis (MST) method has been described in detail previously [14,15,–16]. For an optimal protein to dye (RED-tris-NTA, Nanotemper, Munich, Germany) ratio for subsequent His-tag labeling, MST experiments were conducted in buffer containing 25 mM HEPES (pH 7.4), 150 mM NaCl and 0.05% pluronicTM F-127. In addition, 150 nM RORγt (residues 265–509) protein and 5 nM dye were used for the assay. The compound was serially diluted from ~10 μM to 5 nM. The medium MST power and 11% excitation power were applied to the signal detection of protein in the 25.0 °C thermostat set point. The instrument used in the experiment is monolith NT Automated (Nanotemper, Munich, Germany). Data analyses were performed using Nanotemper Analysis software (MO Affinity Analysis), and curves were plotted in GraphPad Prism.
Protein thermal shift assay (PTS)
In the assay, 7.5 μM purified RORγt-LBD proteins and 5× SYPRO® Orange (Molecular Probes, Life Technologies, Inc., Eugene, OR) were mixed into a fast 96-well optical plate (Applied Biosystems, Foster City, CA, USA) in buffer consisting of 25 mM HEPES (pH 7.4) and 150 mM NaCl. Then, the compounds were diluted to different concentrations by DMSO and added to the mixture at different concentrations. The fluorescence signals were gathered with a slope of 2.5 °C·min−1 from 25 °C to 95 °C using a real-time PCR machine (QuantStudio™ 6 Flex, Applied Biosystems). The output data were analyzed, melting temperatures were calculated by Protein Thermal Shift™ v1.0 software (Life Technologies), and curves were plotted in GraphPad Prism.
Crystallization, data collection, and structure determination
First, his-thrombin RORγt (residues 260–507)-GGG-SRC1 proteins were prepared at 10 mg/mL, and initial crystal trials were conducted utilizing the hanging drop vapor diffusion method in 24-well plates at 25 °C. The apo crystals were grown by mixing 1 μL of the protein (10 mg/mL) with 1 μL of reservoir solution containing 0.4 M NaCl, 8% PEG4000, and 0.1 M PIPES, pH 6.5. The inerratic crystals appeared in 1 day and grew to their full size in ~1 week. The cocrystals of inverse agonist compound 6 were obtained by soaking the compound into apo RORγt-LBD-SRC1 crystal. Nevertheless, crystals of RORγt-LBD-SRC1 in complex with agonist compound 7d and inverse agonist compound 7h were prepared by incubation of 10 mg/mL protein with 0.5 mM compounds. After 5 h of incubation, the cocrystals were obtained fortunately by mixing 1 μL of the protein with 1 μL of reservoir solution containing 0.4 M NaCl, 8% PEG4000, and 0.1 M PIPES, pH 6.5. The diffraction data of compounds 6, 7d, and 7h were collected on beamlines BL17U1 and BL19U1 at Shanghai Synchrotron Radiation Facilities at 100 K [17, 18]. Data sets were processed (indexing and integration) using the program HKL3000 or HKL2000 [19]. The structures were determined by molecular replacement using the programs PHENIX [20]. The models were refined using the program PHENIX and rebuilt with COOT [21]. The stereo images and electron density maps were generated using PyMol.
Model construction and preparation
Initial structures were obtained from our crystal coordinates (PDB code: 6LOA, 6LOB, and 6LO9), including compound 6-RORγt, compound 7d-RORγt and compound 7h-RORγt. For the models of MD simulation, the triglycine linker and cofactor SRC1 were removed from the crystal structures. Each protein structure was prepared by assigning the bond order, adding missing hydrogen, and deleting water molecules, followed by Impref minimization with Maestro (Schrödinger, LLC, NY, USA). For ligand optimization, the Gaussian 09 (Gaussian 09, Revision D. 01, Gaussian, Inc., LA, USA) program was used to calculate the partial atomic charges of ligands (compounds 6, 7d, and 7h) at the HF/6-31 G* level. The restrained electrostatic potential charges of the ligands were derived with an antechamber. The simulation systems were subsequently constructed by means of tleap in AmberTools 16 (AmberTools 16, University of California, CA, USA). Each complex was solvated in a cubic water box of TIP3P water molecules, maintaining a distance of 8 Å from any protein atom. A suitable number of counterions was added to neutralize the charge of each system.
MD simulations
All simulations were performed by Amber 16 with GPU acceleration (AmberTools 16). The ff14SB force field was used for each protein [22], and the general Amber force field was used for each molecule [23]. Each system was minimized with the steepest descent minimization of 5000 steps and a conjugate gradient minimization of 5000 steps by freezing heavy atoms of complex with a harmonic restraint of force constant of 1 kcal/(mol·Å2). Then, each system was heated to 300 K gradually within 0.2 ns. Then, this step was followed by another 0.1 ns equilibrium at 300 K in the constant-pressure, constant-temperature ensemble (NPT). Finally, 1000 ns conventional MD production without any restriction was performed. Particle mesh Ewald was applied to calculate the electrostatic interactions [24]. A cutoff distance of 12 Å was used for nonbonding van der Waals and electrostatic interaction calculations. Data analysis was performed on the trajectories of the MD production stage. The MD trajectory postprocessing and data analysis were implemented inside the cpptraj module in Amber16. The representative structure for each system was derived from the last 400 ns stage, in addition to the initial conformation. All the structure diagrams were processed with PyMOL software (Version 1.2r3pre, Schrödinger).
Results
Identification of the “Short” RORγt inverse agonist
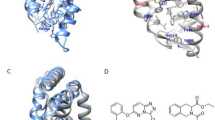
In a FRET assay where a surrogate agonist was added, 6 exhibited RORγt inhibitory activity with an IC50 of 167 nM and a maximum percentage inhibition (% max inhibition) of 153% [13]. In a dual FRET assay without a surrogate agonist added, 6 showed RORγt inhibitory activity with an IC50 of 45.3 nM and a % max inhibition of 71.7%, indicating that it is an inverse agonist. To confirm a direct interaction of RORγt-LBD with compound 6, we first conducted a MST assay and PTS assay. As shown in Supplementary Fig. S1a, compound 6 displayed a Kd value of 637.5 ± 109.6 nM. In addition, compound 6 changed the Tm value (ΔTm) in a concentration-dependent manner, with the ΔTm values of 8.7 °C and 8.1 °C at 25 μM and 7 μM, respectively. Then, inverse agonist 6 was cocrystallized with RORγt LBD (PDB code: 6LOB, resolution of 2.40 Å) to help elucidate the binding mode. Data collection and refinement statistics are included in Supplementary Table S2. With the hydrogen bond between the carboxamide linker and Phe377 and with the sulfonyl group interacting with Leu287 and Arg367 through hydrogen bonds, compound 6 made strong polar interactions at the hydrophilic site. However, the interaction at the AF2 domain was not well established. Although the 6-position of the carbazole-ring directly pointed to AF2, it was ~6.5 Å, 6.7 Å, and 7.1 Å from His479 (center of imidazole ring), Tyr502 (O) and Trp317 (center of benzene ring), respectively, which left a cavity of ~2–3 atoms in size between the 6-position of the carbazole-ring and these key residues (Fig. 3 and Supplementary Fig. S2a).
Left: Surface representation that shows compound 6 (pink) binding to RORγt (blue-white), along with a 2mFo-DFc OMIT electron density map at a contour level of 2.0δ shown as a blue mesh around the compound. Right: Compound 6 forms extensive interactions with the RORγt-LBD. Hydrogen bond interactions are indicated by dashed lines in orange. Distances from C6 of the carbazole ring are marked in green. For clarity, only important residues (blue-white) at the binding site are shown.
These observations indicate that compound 6 is a “short” inverse agonist, which leads to the hypothesis that introducing R group substituents to the 6-position will stabilize or destabilize the AF2 domain to potentially turn the “short” inverse agonist 6 into agonists and then “long” inverse agonists. Therefore, a series of carbazole-based analogs with substituents at the 6-position gradually increasing was designed and synthesized.
From “Short” inverse agonists to agonists
With the substituents growing, inverse agonist 7a first showed a decrease in % max inhibition and a rise in potency (RORγt dual FRET IC50 = 20.6 nM, % max inhibition = 45.5%) compared with 6 (RORγt dual FRET IC50 = 45.3 nM, % max inhibition = 71.7%, Table 1 and Supplementary Table S1). Based on this result, we kept increasing the size of the substituent group on the carbazole ring. As we expected, the first carbazole-based agonist was discovered. A methoxyl group at the 6-position reversed the function and led to agonist 7b with an EC50 of 8.0 nM and a moderate % maximum activation (52.9%, Table 1). Enlarging the substituent by an ethoxyl group led to agonist 7c with a higher efficacy (% max activation = 61.9%, Table 1), and agonist 7d with an isopropoxyl group exhibited the best agonist activity with a potent maximum activation of 83.7%. Interestingly, 7e, 7f and 7g with larger substituents, such as tert-butoxyl, cyclobutoxy, and cyclopentoxy groups, started to show a decrease in efficacy, giving a mitigated % max activation (59.1%, 48.8%, and 42.6%, respectively, Table 1 and Supplementary Table S1) in the dual FRET assay. The functional switch phenomenon observed in the dual FRET assay was confirmed by the cell-based reporter gene assay. In the Gal4 cell-based reporter gene assay, the compounds’ activation percentages at 300 nM ranged from −48.12% (inverse agonist 6) to 48.60% (agonist 7d) and then to a reduced activation level of 13.30% (agonist 7g), which was consistent with our hypothesis that introducing R substituents to approach the AF2 domain will functionally reverse “short” inverse agonists to agonists (Table 1).
Compound 7d was then measured in the MST assay and PTS assay (Supplementary Fig. S1b). The results revealed a higher affinity of 53.3 ± 17.4 nM and a similar ΔTm changing mode compared with those of compound 6. Fortunately, we succeeded in acquiring a cocrystal structure of RORγt-LBD with 7d (PDB code: 6LOA, resolution of 2.51 Å). This structure showed that 7d occupied the RORγt-LBD and fit well in the electron density map. In addition, the cocrystal structures of compounds 6 and 7d overlapped well in both the carbazole moiety and the sulfonyl moiety toward Phe377, Phe378, Phe388, Leu287, and Arg367 (Supplementary Fig. S3). The terminal isopropoxyl group of agonist 7d was observed pointing to the AF2 domain, 4.8 Å and 3.4 Å apart from His479 (center of imidazole ring) and Tyr502(O), packing against the hydrogen bond between His479(NE2) and Tyr502(OH) (Fig. 4a and Supplementary Fig. S2b). In addition, the formation of an alkyl-π interaction of 4.7 Å toward the phenyl ring of Trp317 was found. These results could explain the stabilized AF2 domain and the observed inverse agonist-agonist functional switch.
Crystal structures of RORγt(265-509)-LBD with modulators. a With compound 7d, PDB code: 6LOA, resolution of 2.51 Å. b With compound 7h, PDB code: 6LO9, resolution of 1.85 Å. Left: Surface representation that shows compound binding to RORγt (blue-white), along with a 2mFo-DFc OMIT electron density map at a contour level of 2.0δ shown as a blue mesh around the compound. Right: The compound forms extensive interactions with the RORγt-LBD. Hydrogen bond interactions are indicated by dashed lines in orange. The green lines represent distances.
From agonists to “Long” inverse agonists
However, when the substituents increased to cyclohexyloxy group, the compound started to show a functional switch in the opposite direction. With an IC50 of 88.3 nM and a % max inhibition of 40.7% in the dual FRET assay (Supplementary Table S1), compound 7h functionally changed to a so-called “long” inverse agonist. The inhibitory activity was also confirmed by a moderate-to-potent % max inhibition of 67.4% in the cell-based reporter gene assay. With the substituents growing continuously, the inhibition efficacy of compound 7i increased to 84.3%, and compound 7j functioned as the best “long” inverse agonist with a similar % max inhibition of 94.4% in the dual FRET assay (Table 1). However, in the GAL4 cell-based reporter gene assay, compounds 7i and 7j only displayed inhibitory activities of 16.3% and 28.5%, respectively. In contrast to the results of the “short” ones, the efficacies in the dual FRET assay and cell-based reporter gene assay have some gap. This finding might be explained by the reduced cell permeability according to the QikProp predictions (Supplementary Table S3). Similarly, MST and PTS assays were also conducted with compound 7h to determine its binding ability. A decreased Kd value of 159.4 ± 65.4 nM and ΔTm values of ~8.6 °C and 7.8 °C were obtained (Supplementary Fig. S1c). Notably, we obtained a cocrystal structure of RORγt-LBD with 7h (PDB code: 6LO9, resolution of 1.85 Å) for further molecular mechanistic exploration. The distances between the mass center of the cyclohexyl ring of 7h and the key residues were similar to those in the agonist 7d-RORγt complex. However, the cyclohexyl group of 7h bound in a boat conformation instead of a chair conformation, indicating an existing collision between the 4-position carbon atom (C4) of the cyclohexyl group and the AF2 domain (Fig. 4b and Supplementary Fig. S2c).
With the above cocrystal structures in hand, we superimposed the three structures of 6, 7d, and 7h to compare the overall binding modes. As shown in Fig. 5, His479 and Tyr502 rotate slightly, as indicated by the red arrow in crystal structures. Since the aryl sulfone moiety of ligands 6, 7d, and 7h superimposed well, we surmised that the stabilization of AF2 was affected by the terminal substituents on the carbazole moiety, which made apparent agonist/inverse agonist differences in biological function.
MD simulation study
To understand the role of terminal substituents of ligands in the switching function of RORγt, we conducted a 1 μs MD simulation. As reported previously, the hydrogen-bonding interaction between His479 and Tyr502 played a key role in stabilizing the position of H12 in the active form [25]. Therefore, we first monitored the distance between His479(NE2) and Tyr502(OH) during the MD trajectory (Fig. 6a). In our MD simulation study, no dynamic change in distance was observed (4.17 ± 0.89 Å) in 7d-RORγt, whereas the distances gradually increased after 600 ns in 6-RORγt and 7h-RORγt, which was consistent with the response of the agonist lock between His479 and Tyr502 in RORγt. To determine the dominant reason for the His479-Tyr502 hydrogen bond disruption, we monitored the time-dependent distances between the 6-position carbon atom (C6) of the carbazole scaffold and His479 (center of imidazole ring)/Tyr502(O) (Fig. 6b). In the agonist 7d-RORγt system, the distances between C6 and the residues were quite stable. In the “short” inverse agonist 6-RORγt system, the time-dependent distance curve of C6-Tyr502 exhibited apparent fluctuations during 608–615 ns and 810–814 ns. Less obvious fluctuation was observed during 430–500 and 820–1000 ns, whereas the C6-His479 distance curve remained flat (4.77 ± 0.45 Å) during the entire simulation time. In the “long” inverse agonist 7h-RORγt system, the distance curve of C6-His479 decreased slightly at ~620 ns, followed by a significant fluctuation in the C6-Tyr502 distance during 801–808 ns. This result indicated that the behaviors of His479 and Tyr502 in the three systems were significantly different.
a Time-dependent distances between His479(NE2) and Tyr502(OH), and the cutoff of the hydrogen bond distance is 3.5 Å. b Time-dependent distances between the C6 atom of each ligand and two residues, His479 (center of imidazole ring) and Tyr502(OH). Chi1 and Chi2 angle distribution for His479 (c) and Tyr502 (d) (left three panels). Contours are drawn at <0 (black), 0 (red), 0.5…, 19.5, 20 (blue), and >20 (white) kcal/mol. Representative snapshots of the local energy minima in marked regions (right panel) during simulations. e–g Graphical representation of the conformational states of His479 and Tyr502 observed during MD simulation. e “Short” inverse agonist 6 complexes are shown in the pink series (at 0 ns, 612 ns, 810 ns, and 1000 ns); (f) “Long” inverse agonist 7h complexes are shown in the orange series (at 0 ns, 810 ns, 820 ns, and 1000 ns); (g) Agonist 7d complexes are shown in the green series (at 0 ns and 1000 ns). Conformation snapshots over simulation time are colored in various shades that start with white and fade down to the darker. Each arrow represents the change of the His479 or Tyr502 side chain in direction.
In addition to the time-dependent distance, the side-chain dihedral angle was analyzed to reveal the specific conformational changes of His479 and Tyr502. The distribution of Chi1 and Chi2 during all simulation times is visualized in Fig. 6c, d. It was obvious that dihedral angles appeared with different frequencies, especially the Chi1 angles. Since significant conformational changes in His479 and Tyr502 were detected in the late MD simulation, the following dihedral angle analysis and representative structure presentation would be focused on the last 400 ns stage. For ease of comparison among the three systems, the Chi2 angle was restricted as −110° for His479 and −90° for Tyr502, where the fixed Chi2 angles were determined by the starting systems. The conformation in the lowest energy minima for each system is marked in the left panels, indicating that the His479 Chi1s for 6-RORγt and 7d-RORγt were preferred in the trans position (Chi1s near 175° and 155°, respectively), whereas the His479 Chi1 for 7h-RORγt was preferred in the g- position (Chi1 near −75°). As shown in the right panel of Fig. 6c, the His479 side chain might be crashed by a chair-form cyclohexyl group of 7h and had a deflection away from the ligand. In particular, the g- position for Tyr502 Chi1 was preferred in 6-RORγt, while the g+ position was most frequent in 7h-RORγt (Fig. 6d), which all differed from the trans position represented in 7d-RORγt.
The graphical representation of conformation states of His479 and Tyr502 in H11-H12’-H12 during MD simulations is presented (Fig. 6e-g). In the “short” inverse agonist 6-RORγt system, because of the unoccupied cavity around the 6-position of the carbazole ring, His479 flipped toward the ligand, and the Tyr502 side chain then shifted outwards from the inside pocket (Fig. 6e), which led to the disruption of the His479-Tyr502 hydrogen bond. Not surprisingly, the cyclohexyl group in the “long” inverse agonist 7h clashed with His479, followed by the Tyr502 side-chain flipping away from the pocket (Fig. 6f). Agonist 7d with an isopropoxy group, however, helped to form the His479-Tyr502 hydrogen bond and relatively stabilized the conformation of H12 (Fig. 6g).
Intrigued by the different behaviors of Tyr502 and His479 in “short” and “long” inverse agonist-RORγt systems, we suspected that the conformation of the whole RORγt in the two systems may also show different changes. Therefore, we calculated the root mean square fluctuation for each residue and found that the H11-H11’-H12 portion was highly flexible in both systems (Supplementary Fig. S4). To obtain the individual changing modes of H11, H11’, and H12, we separately calculated the root mean square deviation (RMSD). With “short” inverse agonist 6, H11 heavy atoms remained relatively stable. However, the RMSD of H12 heavy atoms coordinated with the general changing trends of the C6-Tyr502 curve during 0–810 ns and decreased during 810–815 ns. It increased again after 815 ns, together with the H11’ heavy atoms (Fig. 7a). Interestingly, although the behavior of the H11 and H11’ backbones remained consistent with their heavy atoms, the RMSD value of the H12 backbone remained flat during the whole simulation time (Fig. 7c), indicating that the introduction of compound 6 could affect the side chain of H12 during 0–815 ns and the backbone of H11’ after 815 ns. However, in the 7h-RORγt system, the three helixes appeared to be more flexible during 0–650 ns (Figs. 7b, d). H11’ became stable, and the RMSD of H11 heavy atoms increased slowly after 650 ns, 80 ns ahead of the rise in the RMSD of the H11 backbone. After 750 ns, the RMSD of H12 heavy atoms started to increase, followed by an increase in the RMSD of the H12 backbone after 800 ns. It seemed that the “long” inverse agonist 7h could influence the helix structures of H11 and H12 by affecting the side chains.
Since α-helixes were generally stabilized by internal hydrogen bonds between every four amino acids, we then evaluated the stability of helixes H11, H11’, and H12 in the compound 6-RORγt system and compound 7h-RORγt system by tracking the distance between residue X (backbone atom O) and residue X + 4 (backbone atom N). Representative time-dependent distance curves in H11, H11’, and H12 are presented in Fig. 8a. In the 6-RORγt system, although hydrogen bonds in H11 and H12 remained stable during the whole simulation time, the connection between Val493(O) and Ala497(N) in H11’ loosened slowly after 800 ns, followed by the fluctuation of C6-Tyr502 distance during 810–814 ns and a gradual increase of the Pro491(O)-Gln495(N) distance after 815 ns. The results suggested that the increased instability of H11’ in the “short” inverse agonist 6-RORγt system could result in H12 movement as a whole, and the swinging H12 further loosened the secondary structure of H11’ as feedback. In another case of the “long” inverse agonist 7h, the hydrogen bond distances of Arg482(O)-Phe486(N) and Leu483(O)-Gln487(N) in H11 started to increase from 799 ns and 797 ns, respectively, after His479 shifted its location ~620 ns, immediately before a significant increase in the C6-Tyr502 distance (801–808 ns) and an addition in the Pro500(O)-Glu504(N) distance (803–810 ns). Therefore, it was reasonable to conclude that in the “long” inverse agonist 7h-RORγt system, the shift of His479 separated the secondary structure of H11, leading to the fluctuation of Tyr502, and then unwound H12.
a The tracked time-dependent distances for important hydrogen bonds of H11, H11’ and H12 in the compound 6-RORγt system and compound 7h-RORγt system. Arg482-Phe486 stands for the distance from the oxygen atom at the Arg482 backbone to the backbone nitrogen atom of Phe486. Leu483-Gln487, Pro491-Gln495, Val493-Ala497 and Pro500-Glu504 stand for their own distance in the same way. Graphic representations of H11, H11’ and H12 (b: with “short” inverse agonist 6, c: with “long” inverse agonist 7h, white: crystal structure, pink and orange: schematic structures at the end of MD simulations).
On the basis of the evidence in this study, we proposed the following models to describe the different behaviors of RORγt induced by our carbazole-containing “short” and “long” inverse agonists. For the “short” inverse agonist 6, lacking a substituent on the 6-position of the carbazole ring to reach the hydrophobic AF2 domain, His479 first flipped toward the pocket side quickly and then destroyed the His479-Tyr502 hydrogen bond, leading to a loosened H11’ and dislocation of H12. In this process, the main protein scaffolds of H11 and H12 were stable, but the helix structure of H11’ and the side-chain of H12 were affected (Fig. 8b). The “long” inverse agonist 7h, with its cyclohexyl group approaching the AF2 domain, clashed sterically with the His479 side chain. His479 then influenced the subsequent residues Arg482 and Leu483, resulting in the movement of Tyr502 toward the exterior part of the binding pocket, which led to the Tyr502-Pro500 collision and the disconnection between Pro500(O) and Glu504(N). As a result, the helix structures of H11 and H12 were partly destroyed (Fig. 8c). Furthermore, a DSSP analysis was conducted to confirm the models proposed from our observations (Supplementary Fig. S5). We determined that destroying whether the location of H12 by the “short” inverse agonist 6 or the helix structure of H12 by the “long” inverse agonist 7h always affected the action of H12 to recruit coactivators, which would thus interrupt or repress the transcriptional function of RORγt.
Discussion
RORγt inverse agonists and agonists have attracted extensive attention as novel therapeutic molecules for autoimmune diseases and cancer immunotherapy in clinical trials. In previous work, the functional characteristics of agonists and inverse agonists have been elucidated [2, 11]. In our study, we first discovered a series of 6-substituted RORγt modulators, including agonists, “short” and “long” inverse agonists, through insights into the cocrystal structure of RORγt with inverse agonist 6, indicating a potent approach to identify agonists and certain types of inverse agonists. By using MD simulation data, we revealed that agonists stabilize and inverse agonists destroy the hydrogen bond between His479 and Tyr502, which confirmed previous work. Although some reports suggested that the conformation of Trp317 might affect the inverse agonist-agonist functional switch [26], there was no obvious conformational change in Trp317 in our study (Supplementary Fig. S6).
Furthermore, as reported recently [12], “short” inverse agonists and “long” inverse agonists differed in the cofactor recruitment assay. However, the molecular mechanisms between “short” inverse agonists and “long” inverse agonists remain unclear. Herein, starting from the minor difference observed in cocrystal structures, we analyzed the side-chain dihedral angles of His479 and Tyr502 and found that the two residues flipped toward opposite directions when introducing “short” inverse agonist 6 or “long” inverse agonist 7h, indicating a different MOA between “short” or “long” inverse agonists. Further analyses showed that the “long” inverse agonists could change the skeletal structures of H11 and H12, the helixes that were important to recruit coactivators, while the “short” inverse agonists were more likely to break the helix structure of H11’ and to influence side chains of H11 and H12, resulting in the dislocation of H12. To the best of our knowledge, this is the first study to discuss the difference between “short” and “long” inverse agonists by proposing detailed molecular mechanisms.
According to previous reports, inverse agonists with a common scaffold may still significantly differ in gene expression, such as CXCR4 and CTLA4, and may affect RORγt-DNA interactions with different mechanisms [27]. Therefore, more experiments, including ChIP-seq and RNA-seq, are needed to understand their unique regulatory functions. We believe that the crystallography-guided design of functionally switchable RORγt modulators and the models we proposed to analyze the behaviors of agonists, “short” and “long” inverse agonists will be helpful to better understand the RORγt modulators at the molecular level and to make the drug discovery process more efficient.
In conclusion, we designed a series of carbazole-based RORγt modulators using crystallography-guided design and succeeded in the identification of agonists, “short” and “long” inverse agonists in one scaffold. The “short” and “long” inverse agonists were found to behave differently through MD simulation analysis. The different molecular mechanisms of “short” and “long” inverse agonists can be important to the drug discovery process and therefore deserve more experiments to define their regulatory functions in the future.
Accession codes
The coordinates and structural factors have been deposited in the Protein Data Bank with the accession codes 6LOB (6), 6LOA (7d) and 6LO9 (7h). The authors will release the atomic coordinates and experimental data upon article publication.
References
Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33.
Yuan C, Chen H, Sun N, Ma X, Xu J, Fu W. Molecular dynamics simulations on RORgammat: insights into its functional agonism and inverse agonism. Acta Pharmacol Sin. 2019;40:1480–9.
Li X, Anderson M, Collin D, Muegge I, Wan J, Brennan D, et al. Structural studies unravel the active conformation of apo RORgammat nuclear receptor and a common inverse agonism of two diverse classes of RORgammat inhibitors. J Biol Chem. 2017;292:11618–30.
Kallen J, Izaac A, Be C, Arista L, Orain D, Kaupmann K, et al. Structural states of RORgammat: X-ray elucidation of molecular mechanisms and binding interactions for natural and synthetic compounds. ChemMedChem. 2017;12:1014–21.
Mahalingam D, Wang J, Hamilton EP, Sarantopoulos J, Nemunaitis J, Weems G, et al. Phase 1 open-label, multicenter study of first-in-class RORγ agonist LYC-55716 (Cintirorgon): safety, tolerability, and preliminary evidence of antitumor activity. Clin Cancer Res. 2019;25:3508–16.
Sun N, Guo H, Wang Y. Retinoic acid receptor-related orphan receptor gamma-t (RORγt) inhibitors in clinical development for the treatment of autoimmune diseases: a patent review (2016-present). Expert Opin Ther Pat. 2019;29:663–74.
Pandya VB, Kumar S, Sachchidanand, Sharma R, Desai RC. Combating autoimmune diseases with retinoic acid receptor-related orphan receptor-gamma (RORgamma or RORc) inhibitors: hits and misses. J Med Chem. 2018;61:10976–95.
Qiu R, Wang Y. Retinoic acid receptor-related orphan receptor γt (RORγt) agonists as potential small molecule therapeutics for cancer immunotherapy. J Med Chem. 2018;61:5794–804.
Wang Y, Yang T, Liu Q, Ma Y, Yang L, Zhou L, et al. Discovery of N-(4-aryl-5-aryloxy-thiazol-2-yl)-amides as potent RORγt inverse agonists. Bioorgan Med Chem. 2015;23:5293–302.
Rene O, Fauber BP, Boenig GD, Burton B, Eidenschenk C, Everett C, et al. Minor structural change to tertiary sulfonamide RORc ligands led to opposite mechanisms of action. ACS Med Chem Lett. 2015;6:276–81.
Yukawa T, Nara Y, Kono M, Sato A, Oda T, Takagi T, et al. Design, synthesis, and biological evaluation of retinoic acid-related orphan receptor gammat (RORgammat) agonist structure-based functionality switching approach from in house RORgammat inverse agonist to RORgammat agonist. J Med Chem. 2019;62:1167–79.
Wang Y, Cai W, Tang T, Liu Q, Yang T, Yang L, et al. From RORγt agonist to two types of RORγt inverse agonists. ACS Med Chem Lett. 2018;9:120–4.
Huang Y, Yu M, Sun N, Tang T, Yu F, Song X, et al. Discovery of carbazole carboxamides as novel RORγt inverse agonists. Eur J Med Chem. 2018;148:465–76.
Witosch J, Wolf E, Mizuno N. Architecture and ssDNA interaction of the Timeless-Tipin-RPA complex. Nucleic Acids Res. 2014;42:12912–27.
Gaffarogullari EC, Krause A, Balbo J, Herten DP, Jaschke A. Microscale thermophoresis provides insights into mechanism and thermodynamics of ribozyme catalysis. RNA Biol. 2013;10:1815–21.
Parker JL, Newstead S. Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 2014;507:68–72.
Wang Q, Zhang K, Cui Y, Wang Z, Pan Q, Liu K, et al. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl Sci Tech. 2018;29:68.
Zhang W, Tang J, Wang S, Wang Z, Qin W, He J. The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nucl Sci Tech. 2019;30:170.
Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–66.
Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21.
Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501.
Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–713.
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–74.
Harvey MJ, De Fabritiis G. An implementation of the smooth particle Mesh Ewald method on GPU hardware. J Chem Theory Comput. 2009;5:2371–7.
Fauber BP, Magnuson S. Modulators of the nuclear receptor retinoic acid receptor-related orphan receptor-γ (RORγ or RORc). J Med Chem. 2014;57:5871–92.
Sun N, Yuan C, Ma X, Wang Y, Gu X, Fu W. Molecular mechanism of action of RORγt agonists and inverse agonists: insights from molecular dynamics simulation. Molecules. 2018;23:3181.
Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, et al. Small-molecule RORgammat antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 2014;40:477–89.
Acknowledgements
We are extremely grateful to the National Centre for Protein Science Shanghai (Protein Expression and Purification system) for their instrument support and technical assistance. We thank the staff from the BL17U1 and BL19U1 beamlines at the Shanghai Synchrotron Radiation Facility (SSRF) for assistance during data collection. We gratefully acknowledge the financial support from the National Natural Science Foundation of China (Grant Numbers: 81703415; 81573276; 81874287; 81973163), the K. C. Wong Education to CL, the Fudan-SIMM Joint Research Fund (Grant Number: FUSIMM20174007), the CAS Strategic Priority Research Program (Grant Number: XDA12020372), the Shanghai Biopharmaceutical Science and Technology Supporting Plan (Grant Number: 17431902100; 19431900100), the National Science and Technology Major Project (Grant Number: 2018ZX09711002-003-014), the Natural Science Foundation of Shanghai (Grant Number: 19ZR1436700), and the China Postdoctoral Science Foundation (Grant Number: 2019M651383).
Author information
Authors and Affiliations
Contributions
MCY, FY, XYD, NNS, QX, HLJ, KXC, CL, XML, SJC, and YHW developed the hypothesis, designed the experiments, and revised the manuscript. MCY, ZYJ, YFH, YRY, and CZ designed and synthesized the compounds. FY, ZFC, and SQG performed the crystallographic studies. MCY, XYD, and NNS conducted the MD simulation and data analysis. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Yu, Mc., Yang, F., Ding, Xy. et al. Crystallography-guided discovery of carbazole-based retinoic acid-related orphan receptor gamma-t (RORγt) modulators: insights into different protein behaviors with “short” and “long” inverse agonists. Acta Pharmacol Sin 42, 1524–1534 (2021). https://doi.org/10.1038/s41401-020-00552-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-020-00552-w