Abstract
Adolescent individuals exhibit great variability in cortical dynamics and behavioral outcomes. The developing adolescent brain is highly sensitive to social experiences and environmental insults, influencing how personality traits emerge. A distinct pattern of mitochondrial gene expression in the prefrontal cortex (PFC) during adolescence underscores the essential role of mitochondria in brain maturation and the development of mental illnesses. Mitochondrial features in certain brain regions account for behavioral differences in adulthood. However, it remains unclear whether distinct adolescent behavioral phenotypes and the behavioral consequences of early adolescent stress exposure in rats are accompanied by changes in PFC mitochondria-related genes and mitochondria respiratory chain capacity. We performed a behavioral characterization during late adolescence (postnatal day, PND 47–50), including naïve animals and a group exposed to stress from PND 31–40 (10 days of footshock and 3 restraint sessions) by z-normalized data from three behavioral domains: anxiety (light–dark box tests), sociability (social interaction test) and cognition (novel-object recognition test). Employing principal component analysis, we identified three clusters: naïve with higher-behavioral z-score (HBZ), naïve with lower-behavioral z-score (LBZ), and stressed animals. Genome-wide transcriptional profiling unveiled differences in the expression of mitochondria-related genes in both naïve LBZ and stressed animals compared to naïve HBZ. Genes encoding subunits of oxidative phosphorylation complexes were significantly down-regulated in both naïve LBZ and stressed animals and positively correlated with behavioral z-score of phenotypes. Our network topology analysis of mitochondria-associated genes found Ndufa10 and Cox6a1 genes as central identifiers for naïve LBZ and stressed animals, respectively. Through high-resolution respirometry analysis, we found that both naïve LBZ and stressed animals exhibited a reduced prefrontal phosphorylation capacity and redox dysregulation. Our findings identify an association between mitochondrial features and distinct adolescent behavioral phenotypes while also underscoring the detrimental functional consequences of adolescent stress on the PFC.
Similar content being viewed by others
Introduction
Adolescence is a period of learning, social adaptation, and dynamic neurobiological maturation, which begins with puberty and ends in adulthood [1]. Evident structural and functional alterations occur in the brain during adolescence, particularly within cognitive [2], social [3], and emotional processes [4]. These changes are particularly pronounced in cortical regions: the latest brain areas to mature [5]. Throughout adolescence, functional connectivity is strengthened between cortical regions and limbic circuits [6]. This leads to better stabilization and efficiency of prefrontal cortex (PFC) activity [7], a crucial component for higher-order cognitive processes [8]. PFC maturation also reduces inter-individual variability in cognitive task performance and enhances overall brain function, refining complex behaviors [9].
Social and neurobiological factors collectively shape brain development and personality traits [10]. Social experiences influence behavioral outcomes and predict vulnerability and resilience to stress [11]. Adolescence represents a critical period for experience-dependent plasticity in social behaviors [12, 13]. Adolescent animals display significant variability in cortical dynamics [14], which affects behavioral phenotypes related to motivation [15], learning [16], and decision-making [16, 17].
Susceptibility to adverse socio-environmental factors, such as trauma, psychosocial insults, and maltreatment, is also increased in adolescence. Animal studies show the potential of stress during the peripubertal period to cause changes in the brain and behavior in adulthood [17,18,19,20,21,22,23]. Furthermore, the timing of stress seems critical for the behavioral outcomes [24,25,26].
Cortical gene signature during adolescence highlighted increased expression of genes associated with energy metabolism and oxidative phosphorylation (OXPHOS) [27], supporting the role of mitochondria in normal cortical development and behavior [28, 29]. Mitochondria are emerging as a critical regulator of social behaviors and trait anxiety [30,31,32,33]. Changes in mitochondrial function and gene expression are implicated in the modulation of behaviors in response to stress [34]. Therefore, changes in mitochondrial gene expression during adolescence may influence mitochondrial function and redox regulation, potentially leading to behavioral alterations.
Here, we explored the behavioral consequences of stress during late adolescence in rats and linked these outcomes to cortical transcriptomic changes. We first investigated individual phenotype variations within three behavioral domains: anxiety, sociability, and cognition. Then, we performed genome-wide bulk RNA sequencing in the PFC and examined the relationship between mitochondria-related genes and behavioral phenotypes in both naïve and stressed animals. Lastly, we investigated PFC mitochondria respiratory function and reactive oxygen species (ROS) production. Altogether, our findings underscore that differences in mitochondrial gene expression and respiratory function in PFC contribute to individual behavioral variability in naïve animals and highlight the detrimental functional consequences of stress on prefrontal redox regulation.
Materials and methods
Animals
Male and female Sprague-Dawley rats (postnatal day, PND70), an outbred laboratory rat population, were obtained from the Central Animal Facility of the University of São Paulo, Ribeirão Preto, and allowed to acclimatize for one week in the local animal facility before breeding. Mating was confirmed by spermatozoa presence in the vaginal smear, and birthday was defined as PND0. Pups were weaned on PND21. We use only the male offspring, as female adolescent rats do not show the behavioral and electrophysiological changes induced by this stress protocol applied from PND31 to 40 [35, 36]. Animals were randomly assigned to experimental groups, each cage devoted to a specific experimental procedure. Rats were housed (2–3 animals per cage) at temperature- (22 °C) and humidity-controlled (47%) room. The Ribeirão Preto Medical School Ethics Committee (CEUA-FMRP 248/2019) approved the procedures following Brazilian and international regulations.
Adolescent stress protocol
Animals were exposed to inescapable foot shock (FS; from PND31 to 40) daily and three restraint stress (RS) sessions (PND31, 32, and 40), as previously described [24, 26, 37]. Briefly, rats were exposed to one session of FS per day for 10 consecutive days. In each session, animals were placed in a Plexiglas chamber with a grid floor of 0.48 cm stainless steel rods spaced 1.6 cm apart (EP107R, Insight Equipment, Brazil). 25 FS (1.0 mA, 2 s) was delivered pseudo-randomly (5 cycles of 30, 60, 40, 60, and 90 s). Immediately after receiving FS, rats were submitted to RS for 1 h in Plexiglas cylindrical restraint tubes (20.3 × 5.1 cm) ventilated by holes (1 cm diameter) on the first, second, and last day of FS exposure. Naïve animals were left undisturbed in their home cages (n = 18), while stressed animals (n = 14) were subjected to the stress protocol.
Behavioral characterization
One week after stress, rats were tested in the following behavioral tests: light-dark box (LDB) on PND47 to assess anxiety-like behavior; social interaction test (SIT) on PND48 to measure sociability; and novel-object recognition test (NOR) on PND49 to evaluate cognitive function.
LDB
The LDB apparatus has two compartments (240 cm × 210 cm) connected by a door. The light and the dark compartments have a grid floor. Two hours after the EPM, animals were placed in the dark compartment and allowed to explore the apparatus freely for 5 min after the first entry into the light zone. The ANY-maze Software analyzed the time spent in the light zone.
SIT
Animals were placed in the center of a circular arena (D60 cm × H65 cm) and allowed to explore for 5 min. Then, an unfamiliar male Sprague-Dawley rat (PND50–54) was placed in the arena as a social target for 10 min. The social interaction time was measured when the tested animal was sniffing the unfamiliar rat’s anogenital region, head, or body and when they were following, crawling over and under each other. A blind experimenter quantified the social interaction time.
NOR
Two hours after the SIT, each animal was habituated in a circular arena (D60 cm × H60 cm) for 10 min. The NOR test was conducted in the same circular arena 24 h later. Animals were subjected to two trials separated by 1 h. During the first trial (acquisition trial, T1), rats were placed in the arena containing two identical objects for 5 min. For the second trial (retention trial, T2), one of the objects presented in T1 was replaced by an unknown (novel) object. Animals were then placed back in the arena for 5 min. Object exploration was defined as when the animal faced the object at 2 cm of distance or less while watching, licking, sniffing, or touching it with the forepaws while sniffing. A blind experimenter quantified object exploration. Recognition memory was assessed using the discrimination index:
\(\left[\left({{t}}_{{\rm{novel}}\; {\rm{object}}}{{\mbox{-}}}{{t}}_{{\rm{familiar}}\; {\rm{object}}}\right)/\left({{t}}_{{\rm{novel}}\; {\rm{object}}}+{{t}}_{{\rm{familiar}}\; {\rm{object}}}\right)\right]\)
Behavioral z-score index
The integrated behavioral z-score method allows the normalization of individual data observed in different behavioral parameters to decrease the intrinsic variability of single tests and enhance the sensitivity and reliability of the individual phenotyping [38]. We calculated the integrated behavioral z-score by averaging the z-normalized data [(x−mean of naïve group)/(standard deviation of naïve group)] for time spent in light zone (LBD), social interaction time (SIT) and novel object discrimination index (NOR).
Gene expression profiling from the rat PFC
RNA isolation
Following the behavioral tests on PND 51, animals were anesthetized (urethane 25%, 1 mL/100 g/rat) and perfused with cold 0.01 M phosphate-buffered saline (PBS, pH = 7.4). PFC from both hemispheres was collected and snapped frozen in liquid nitrogen until use for RNA extraction (n = 8/group). To this end, we used RNAqueous-Micro Total RNA Isolation Kit (Thermo Fisher Scientific; #AM1931), according to the manufacturer’s instructions.
Bulk RNA-sequencing
The extracted RNA was used for performing the transcriptomic analysis from the PFC of naïve and adolescent-stressed rats using bulk RNA barcoding and sequencing, as previously described [39, 40]. Briefly, the RNA samples were reverse transcribed with individual barcoded oligo-dT primers. Then, all samples were pooled together, and the second strand synthesis generated the double-stranded cDNA via the nick translation method. Illumina-compatible libraries were prepared by tag-mentation of 5 ng of full-length double-stranded cDNA. Then, the library was amplified, profiled, and sequenced using the Illumina NextSeq 500 platform.
Transcriptomic analysis
Following a quality assessment with FastQC [41], gene reads were mapped with HISAT2 onto Rnor_6.0/rn6 genome assembly for Rattus norvegicus [42]. Mapped reads were counted for each gene locus using the featureCounts function of the subread (2.0.2) package [43]. Read counts are available at https://doi.org/10.6084/m9.figshare.24125793.v1. We normalized count data by size factor and applied a variance stabilizing transformation for visualization purposes [44]. Low-abundance genes were removed before data normalization, keeping only genes with at least ten reads in all samples. Subsequently, we performed a generalized linear model to assess differentially expressed genes (DEGs) using the DESeq2 package [45]. p-values were corrected for multiple testing using the Benjamini–Hochberg method [46]. Transcriptomic analysis was performed in R (R Core Team, 2014).
Functional gene set enrichment analysis
Only DEGs with a padj < 0.1 were explored for enriched gene sets and function. Gene set enrichment analyses were derived from bioinformatics resource systems (web servers) for functional annotation and enrichment analyses of gene lists. Database for Annotation, Visualization and Integrated Discovery (DAVID) [47] was used for gene ontology enrichments, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [48] was assayed to examine gene functions, linking genomic information with higher-order functional processes and diseases.
Network visualization and analysis
The transcriptomic data of DEGs were mapped onto unbiased interaction networks using Cytoscape [49, 50]. The STRING app [51] was used to retrieve information about gene–gene associations and subcellular compartments where gene products are located (COMPARTMENTS database) [52]. The STRING database systematically collects and integrates physical and functional protein–protein interactions from various sources, including automated text mining of scientific literature, computational interaction predictions derived from co-expression and conserved genomic context, databases of interaction experiments, and known complexes/pathways from curated sources [51].
We constructed interactive networks, featuring nodes corresponding to DEGs and edges representing predicted functional and/or physical interactions with a confidence score cut-off of 0.5. Then, to create mitochondria-associated gene networks [52], nodes were filtered based on the likelihood of the gene’s association with the mitochondrial compartment with a confidence score ranging from 4.0 to 5.0. To better visualize up and down-regulated genes, nodes were colored according to the quantitative expression data (Log2FoldChange). Nodes with the highest betweenness and closeness centrality for each comparison were identified using the CytoNCA plugin.
Biomolecular analyses
High-resolution respirometry
2 mg of fresh PFC samples (n = 5 naïve HBZ; 5 naïve LBZ; 6 stressed), finely cut into pieces, were permeabilized in BIOPS solution (2.7 mM EGTA, 20 mM imidazole, 20 mM taurine, 50 mM acid 2-(N-morpholino) ethanesulfonic potassium, 0.5 mM dithiothreitol, 6.5 mM MgCl2, 15 mM phosphocreatine, 0.57 mM ATP, pH 7.1) containing 0.01% saponin for 5 min at 4 °C, then carefully transferred to the chambers of a Oxygraph-2k respirometer (Oroboros Instruments, Austria), containing 2 mL of air-saturated respiration medium MIR05 (0.5 mM EGTA, 3 mM MgCl2, 60 mM K-lactobionate, 20 mM taurine, 10 mM KH2PO4, 20 mM HEPES, 110 mM sucrose, 1 g/L albumin, pH 7.1). First, 9 mM glutamate, 5 mM malate, and 10 mM pyruvate were added to the chambers to determine LEAK respiration. Then, OXPHOS capacity linked to complex I activity (CI) was determined after adding 1 mM ADP to the chambers, followed by adding 10 mM. Maximal capacity rates of the electron transfer system (ETS) were defined after 1 pulse of the mitochondrial uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP, 1 μM). Finally, residual oxygen consumption (Rox) rates were determined by adding 1 mM NaCN. Rox rates were subtracted from all other measurements. The experiments were made in duplicate. The O2 flux obtained in each step of the protocol was normalized by the wet weight of the tissue sample used for the analysis.
Amplex™Red assay
Naïve and stressed animals (n = 5 naïve HBZ; 5 naïve LBZ; 6 stressed) were anesthetized (urethane 25%, 1 mL/100 g), and decapitated. PFC tissues were immediately collected and cut into ~1 mm3 cubes. Samples were carefully transferred to cuvette chambers (Hitachi 4500 Fluorescence Spectrophotometer, Japan) with 2 mL of air-saturated respiration medium containing MIR05, 50 μM Amplex™Red reagent, 0.1 U/mL horseradish peroxidase, substrates (9 mM glutamate, 5 mM malate, and 10 mM pyruvate) and 1 mM ADP to determine hydrogen peroxide production by OXPHOS linked to CI. The fluorescence of each step was measured for 100 s. Then, hydrogen peroxide levels were determined as the angular coefficient of the linear regression lines made for the spectrophotometric data. The total protein amount of each sample was measured by Bradford protein assay and used to normalize data.
Statistical analyses
Data were presented as mean ± SEM. The Bartlett’s and Shapiro–Wilk tests were applied to check for variance and normal distribution before the parametric analysis (two-sided Student’s t-test or one-way ANOVA followed by posthoc Turkey’s test). Significant differences were indicated by p < 0.05. In RNA-seq analysis, p-values were corrected using the Benjamini–Hochberg method for multiple testing. A Pearson correlation analysis tested the correlation between integrated behavioral z-score and normalized read counts. Sample size was based on previous works of the research group.
Behavioral data analysis
Each animal’s behavioral z-scores (LDB, SIT, and NOR) were calculated, and their dimensionality was reduced by principal component analysis (PCA) (code available: https://uc-r.github.io/pca). We performed an unsupervised clustering method (k-means) that separated the animals into three clusters in two PC dimensions. We performed the Elbow, Silhouette and Gap statistic methods to confirm the optimal number of clusters (code available: https://uc-r.github.io/kmeans_clustering). Behavioral data analysis was performed in R (R Core Team, 2014).
Results
Adolescent behavioral phenotypes are affected by stress exposure
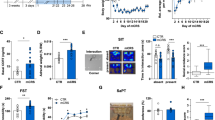
First, we aimed to characterize the adolescent behavioral phenotypes and the impact of stress. Adolescent rats were exposed to the stress protocol (PND31–40) and subsequent anxiety, sociability, and cognitive function behavioral assessments (PND47–49), followed by PFC collection (PND51) (Fig. 1A). Stressed animals exhibited reduced time in the light zone of the LDB, indicating an anxiogenic response (Fig. 1B), and social interaction deficits (Fig. 1C). Moreover, stressed animals showed a decreased novel object recognition index in NOR test, suggesting cognitive impairments (Fig. 1D).
A Experimental timeline: adolescent rats were stressed via inescapable foot shocks from PND 31 to PND 40. From PND 47 to PND 49, animals were tested in the LDB, SIT, and NOR (n = 18 naïve and 14 stressed). Brain tissue was collected on PND 51. Adolescent stress decreased B time spent in the light zone of the LDB (t30 = 5.53, p < 0.0001), C social interaction time (t30 = 7.86, p < 0.0001), and D the discrimination index in the NOR test (t30 = 7.83, p < 0.0001). E Integrated z-score computed from LDB, SIT, and NOR z-scores and F a heatmap of each behavioral z-score indicates the impact of adolescent stress on behavior. Data are shown as mean ± SEM. *p < 0.05; unpaired t-test.
Next, we applied a z-normalization for all behavioral parameters to combine data from all tests into a single index. Notably, the integrated behavioral z-scores (Fig. 1E) and the heatmap displaying z-normalized behavioral data (Fig. 1F) indicate a high intra-group variability among naïve animals. Conversely, stressed animals are narrowly distributed, indicating a homogenous detrimental impact of stress.
Revealing individual behavioral variability
To gain deeper insight into behavioral variability in naïve and stressed animals, we performed a principal component analysis (PCA) on the z-normalized behavioral scores. The clustering analysis, based on the first and second principal components (PC), categorized animals into three distinct groups (Fig. 2A). The highest contributor to the PC1, which accounted for 85.6% of the explained variance, was social interaction time (39.8%), while for PC2, explaining 9.5% of the variance, was predominantly composed by the discrimination index in the NOR test (60.9%) (Fig. 2B). Clusters 1 and 2 comprised naïve animals, while cluster 3 exclusively consisted of stressed animals, being all separated by the contribution of PC1. We refer to these three groups as “naïve with higher-behavioral z-score” (naïve HBZ; cluster 1), “naïve with lower-behavioral z-score” (naïve LBZ; cluster 2), and “stressed animals” (cluster 3), according to their distinct behavioral phenotypes, as revealed by the integrated behavioral z-score) (Fig. 2C). Our findings were validated by the Silhouette, Elbow, and GAP statistical methods (Supplementary Fig. 1A–C), which indicate k = 3 as the optimal number of clusters and supported our PCA and unsupervised k-mean clustering analysis (Supplementary Fig. 1D).
A Expression of significant PCs after behavioral analyses. Dots represent integrated z-scores from the four behavioral paradigms for each rat. Three groups of individuals (gray, blue, and purple dots) were separated by unsupervised k-meaning clustering (n = 9 naïve HBZ, n = 9 naïve LBZ, n = 14 stressed). B Representative contributions of each behavioral paradigm to the two PCA components. C Intergroup comparisons of the integrated behavioral z-score: naïve HBZ, naïve LBZ, and stressed animals (F2,29 = 317.6, p < 0.0001). D Naïve LBZ and stressed animals spent less time in the light zone of the LDB test (F2,29 = 31.32, p < 0.0001), and E presented reduced sociability (F2,29 = 62.71, p < 0.0001). Also, F both groups displayed a decreased discrimination index in the NOR test (F2,29 = 87.83, p < 0.0001). Data are shown as mean ± SEM. *p < 0.05; one-way ANOVA, followed by Tukey’s multiple comparison test.
Compared to naïve HBZ, naïve LBZ showed decreased time spent in the light zone of the LDB (Fig. 2D), reduced sociability (Fig. 2E), and deficits in the NOR test (Fig. 2F). Stressed animals showed marked impairments in all behavioral tests, distinguishing them from both naïve groups (Fig. 2D–F). These data confirm that adolescent naïve animals cluster into two distinct behavioral phenotypes, while adolescent stress exposure impacts all animals, forming a third cluster.
Transcriptomic analysis of the PFC reveals changes in mitochondrial pathways
Considering the variable behavioral phenotypes among naïve animals and their modulation by stress, we postulated that these variations would be reflected in prefrontal gene expression profiles. Employing a hypothesis-free transcriptomic analysis of the PFC, we performed a series of comparisons, contrasting two groups at a time (naïve LBZ vs. naïve HBZ; stressed vs. naïve HBZ; stressed vs. naïve LBZ) (Supplementary Table 1). These analyses revealed 126 DEGs in naïve LBZ (Fig. 3A), 73 DEGs in stressed animals compared to naïve HBZ (Fig. 3B), and 7 DEGs in stressed animals compared to naïve LBZ (Fig. 3C).
Heatmap of normalized mRNA expression levels of differentially expressed genes (DEGs, padj < 0.1) in the PFC comparing A naïve LBZ vs. naïve HBZ, B naïve HBZ vs. stressed animals, and C naïve LBZ vs. stressed animals. The overall numbers of DEGs up and down-regulated are described in their respective heatmaps. The dendrograms represent the hierarchical clustering of genes according to gene expression (Euclidean distance method). D Gene set enrichment analyses of DEGs in the PFC of naïve LBZ and E stressed animals compared to naïve HBZ. Mitochondria-associated pathways are highlighted in black font.
Next, we performed a gene set enrichment analysis to identify the most significantly affected pathways when comparing naïve LBZ and stressed animals to HBZ rats (Fig. 3D, E). At the transcriptomic level, some of the top 10 genes affected common pathways in these groups were associated with mitochondrial function, including “oxidative phosphorylation” and “mitochondrion organization”. The cellular component analysis also revealed “mitochondrion” as an enriched set in both naïve LBZ and stressed animals (Supplementary Fig. 2A, B). Notably, naïve LBZ animals displayed enrichment of gene sets related to “Thermogenesis”. In contrast, stressed animals showed enrichment in “Response to reactive oxygen species” (Fig. 3E). These findings reveal transcriptional disparities in mitochondrial and metabolic genes among behaviorally distinct subgroups.
Mitochondrial-related gene signatures differ between naïve LBZ and stressed animals
Variability in mitochondrial phenotypes within brain regions associated with anxiety, sociability, and cognition has been described in adult outbred [30, 31, 53, 54] and inbred rodents [55]. Therefore, prefrontal DEGs were filtered by their probability to be present in the mitochondrion compartment with a high confidence score (Fig. 4A–C). We identified 15 mitochondria-related genes comparing naïve LBZ and naïve HBZ, with most of them being down-regulated in naïve LBZ, except for Slc1a3 (Fig. 4A). Comparing stressed and naïve HBZ animals, we found 10 mitochondria-related genes, all down-regulated (Fig. 4B). Only one gene (down-regulated Nlrx1) emerged when comparing stressed with naïve LBZ animals (Fig. 4C).
Summary of DEGs maintained by Cytoscape mitochondrion-node filter from A naïve LBZ vs. naïve HBZ, B stressed animals vs. naïve HBZ, and C stressed animals vs. naïve LBZ. The node’s colors represent the corresponding genes’ relative expression (log2(Fold Change)). Nodes rounded with red lines correspond to central identifiers in the network topology analysis. Nodes rounded with black lines correspond to genes from different OXPHOS complexes and were used to correlate the normalized gene expression levels with the integrated behavioral z-score and distinguish gene expression profiles among intergroups. Lines (edges) represent predicted gene–gene interaction. D–G A positive Pearson correlation was observed between the expression levels of OXPHOS genes and behavioral phenotype. r and p values are described in the graphs. Levels of gene expression from different OXPHOS complexes were significantly different among groups: D Complex I = Ndufaf1 (F2,13 = 6.16, p = 0.01), Ndufa10 (F2,13 = 5.63, p = 0.02), and Ndufa11 (F2,13 = 11.80, p = 0.001). E Complex III = Uqcrq (F2,13 = 14.70, p = 0.0005). F Complex IV = Cox6a1 (F2,13 = 25, p < 0.0001), Cox7c (F2,13 = 11, p = 0.001), and Coa8 (F2,13 = 19.4, p = 0.0001). G Complex V = Atp5g1 (F2,13 = 7.21, p = 0.008). Data are shown as mean ± SEM. *p < 0.05; One-way ANOVA, followed by Tukey’s multiple comparison test.
Next, we sought to enable the inference of molecular function associations through an interaction prediction pipeline [51], thereby supporting a more comprehensive interpretation of mitochondria-related gene variation among behavioral clusters [56, 57]. To achieve this, we constructed interactive networks, where nodes represented DEGs with a padj of <0.1, and edges represented predicted functional and/or physical gene–gene interactions (Supplementary Fig. 3A, B). In the network topology analysis of mitochondrial-related genes (Fig. 4A), Ndufa10 was identified as having the highest betweenness and closeness centrality for naïve LBZ. Ndufa10 was associated with the following genes: (1) Ndufa1, Cox6a1, and Atp5g1—subunits of OXPHOS; (2) Cisd1—a member of the CDGSH domain-containing family; and (3) Etfb—an enzyme located in mitochondrial matrix space.
The mitochondrial-related gene with the highest betweenness and closeness centrality for stressed animals was Cox6a1 (Fig. 4B), which was predicted to link with genes encoding (1) subunits of OXPHOS—Ndufa11, Uqcrq, Cox7c, and Atp5g1; (2) a component of MICOS complex—Chchd6; and (3) a ribosomal protein—Rps3. Additionally, stressed animals displayed a second network formed by the interaction of Tomm20 and Fis1 genes. These findings point to distinct molecular functions associated with changes in OXPHOS according to behavioral subgroups.
Expression profiles of mitochondria-related genes in the PFC are associated with behavioral phenotypes
Since “oxidative phosphorylation” was a commonly enriched pathway among subgroups, and most of the mitochondria-related DEGs were directly linked to OXPHOS, we performed an in-depth analysis of gene reads from each electron transport chain complex. Positive correlations were observed for genes of complex I (Ndufaf1, Ndufa10l1 and Ndufa11; Fig. 4D), complex III (Uqcrq; Fig. 4E), complex IV (Cox6a1, Cox7c, and Coa8; Fig. 4F), and complex V (Atp5g1; Fig. 4G) with the behavioral z-scores. Additionally, we re-evaluated gene expression levels based on behavioral phenotypes (Fig. 4D–G). DEGs were significantly down-regulated in both naïve LBZ and stressed animals compared to naïve HBZ, except for Uqcrq and Cox7c (Fig. 4F), which were reduced only in stressed animals. Re-evaluating gene expression levels based on behavioral phenotypes revealed differences in Cox6a1 among groups, with stressed animals being the most affected. Our results support the notion that distinct behavioral phenotypes are correlated with prefrontal mitochondria-related gene expression.
Changes in mitochondrial respiratory function and ROS production rate account for individual behavioral variations and long-lasting stress response
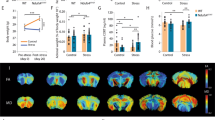
Our transcriptomic analysis alluded to a potential alteration in mitochondrial respiratory function in the PFC of naïve LBZ and stressed animals due to the down-regulation of OXPHOS genes. To explore this possibility, we performed high-resolution respirometry of PFC samples on PND51. Both naïve LBZ and stressed animals exhibited reduced CI and ETS, but not in leak, suggesting a diminished mitochondrial OXPHOS capacity primarily linked to complex I (Fig. 5A). Notably, the integrated behavioral z-score positively correlated with CI (Fig. 5B). Mitochondrial electron transport is the primary source of ROS, and its dysregulation can lead to changes in ROS levels. Accordingly, we found a decrease in hydrogen peroxide production rate in naïve LBZ and stressed animals after stimulating CI (Fig. 5C), which was also associated with behavioral performance (Fig. 5D).
Mitochondrial respiratory capacity and hydrogen peroxide production in the PFC of naïve HBZ, naïve LBZ, and stressed animals was evaluated on PND 51 (n = 5 naïve HBZ, 5 naïve LBZ, and 6 stressed) after behavioral tests. A Naïve LBZ and adolescent stressed animals showed reduced complex I activity (CI) (F2,13 = 27.82, p < 0.0001) and maximal electron transport system capacity (ETS) (F2,13 = 33.95, p < 0.0001), without changing Leak. B A positive Pearson correlation was observed between behavioral phenotypes and OXPHOS respiration linked to CI. C Naïve LBZ and adolescent stress also showed reduced hydrogen peroxide (H2O2) released by CI on PND 51 (F2,13 = 4.43, p = 0.0341). D Positive Pearson correlation with behavioral phenotypes and H2O2 production rate. Data are shown as mean ± SEM. *p < 0.05. One-way ANOVA, followed by Tukey’s multiple comparison test. r and p values are described in the graphs.
Mitochondrial respiratory capacity and hydrogen peroxide production rate are enhanced one day after adolescent stress
Our results pointed to long-lasting changes in prefrontal mitochondrial features in stressed animals, characterized by the down-regulation of OXPHOS genes, enriched pathways related to “response to oxidative stress” and lower complex I respiratory activity 10 days after stress exposure. The temporal changes in mitochondrial respiratory function and ROS abnormal levels after 10 days of stress could represent responses to the “allostatic load” induced by the chronic stress protocol [58, 59]. Then, we postulated that differences in mitochondrial adaptive response under stressful conditions may explain these findings. In another cohort, one day after the adolescent stress (PND41), animals showed higher oxygen consumption at the OXPHOS state linked to complex I activity (Fig. 6A) compared to naïve animals. This aligns with the increased hydrogen peroxide production rate (Fig. 6B).
Discussion
Given the significance of successful adolescent development for adequate sociability and cognitive performance in adulthood, the identification of critical genes moderating distinct behavioral phenotypes and stress responses can shed light on the mechanisms underpinning the development of psychiatric disorders. Here, using adolescent rats, we showed that: (1) naïve animals exhibited considerable behavioral variability, resulting in two distinct behavioral phenotypes and the exposure to adolescent stress led to marked behavioral changes, such as anxiety, reduced sociability, and cognitive impairment; (2) differences in mitochondria-related genes in the PFC correlated with these behavioral phenotypes, with most of the DEGs being down-regulated in both naïve LBZ and stressed animals compared to naïve HBZ; (3) both naïve LBZ and stressed animals showed reduced mitochondrial respiratory capacity and redox dysregulation in the PFC. These findings unveil new molecular-based insights and strengthen the view that brain mitochondria-related genes and mitochondrial respiratory capacity relate to differences in anxiety, sociability, and cognition functions [39, 60,61,62,63].
While individual differences in anxiety traits have been observed in both inbred and outbred adult rodents [30, 31, 53, 54, 64,65,66,67], few studies have investigated this variability in adolescent naïve animals, as in the outbred Sprague Dawley population. Most reports have focused on individual variability in risk-taking, learning, and other cognitive processes during adolescence [8]. In our study, the time spent in the light zone of LDB identified two distinct behavioral profiles in naïve animals and the drastic impact of stress during adolescence. Animals exposed to stress also presented marked deficits in sociability and novel object recognition memory. Inter-individual differences in naïve rats were also observed for these domains. Adolescence is an adaptive period that shapes social and behavioral phenotypes based on individual differences, some insensitive to social contexts [68]. Social behaviors acquired during childhood and adolescence, such as playing and dominance, are crucial for social organization, which impact subsequent interactions with conspecifics and can predict behavioral phenotypes and cognitive skills development [69, 70].
In agreement with reports showing inter-individual variation in sociability among adolescent animals [70, 71], our PCA results defined a PC1 with a significant contribution from the “social interaction time” variable. At the same time, PC2 was influenced by the “novel object recognition index” variable. It is worth noting that when these variables were projected onto a coordinate system, PC1 signs were all negative for cluster 3 (stressed animals), positive for cluster 1 (naïve HBZ), and intermediate to cluster 2 (naïve LBZ). These findings indicate that variability among naïve and stressed animals appears more prominently associated with sociability, even though subgroups also differ in anxiety-like behavior and cognitive function. Indeed, individual differences in anxiety traits are proposed to influence social dominance [30, 66], motivation [31, 66], spatial learning/memory [67], and depressed-like behaviors [31].
Consistent with the idea that mitochondrial features are linked to variability in social behaviors [30,31,32] and are modulated by stress [72], we observed that animals with lower behavioral scores (naïve LBZ and stressed animals) exhibit, except for complex II, down-regulated expression of genes directly linked to OXPHOS, which were positively associated with behavioral performance. Previous studies identified down-regulated genes for several complex I subunits in the dorsal lateral PFC and anterior cingulate cortex in SCZ and autism subjects [73,74,75]. Reduced expression of the Uqcrq gene, which encodes a complex III subunit, was observed only in stressed animals. Genes for complex III subunits are less expressed in cortical regions of patients with neurodevelopmental disorders [75,76,77]. Expression of the gene for complex IV subunit, Cox6a1, was impacted in naïve LBZ and stressed animals compared to naïve HBZ. Moreover, lowered expression levels of complex IV subunits were linked to cognitive impairments in SCZ [78,79,80].
The most down-regulated mitochondrial-related gene in Naïve LBZ was Slc25a12, which encodes the Aralar/slc25a12/AGC1 protein, involved in the transport of aspartate from mitochondria to cytosol in exchange for glutamate. Therefore, it is also a component of the malate-aspartate shuttle, essential to maintain glycolytic pyruvate supply to neuronal mitochondria [81]. A recent study has identified Aralar/slc25a12/AGC1 as the GABA sequesters mitochondrial transporter upon increased mitochondrial activity in Drosophila mutants of the human homolog CYFIP1, leading to social behavioral deficits [82]. Particularly, the only mitochondria-related gene up-regulated in LBZ animals was Slc1a3, which encoded a sodium-dependent, high-affinity amino acid transporter that mediates the uptake of l-glutamate, l-aspartate, and d-aspartate. The transporter, also called glutamate aspartate transporter 1 (GLAST-1), is highly expressed in astrocytes and found in the mitochondrial inner membrane as part of the malate-aspartate shuttle [83]. Metabolite levels for glutamate, glutamine, and GABA in some brain regions, such as the nucleus accumbens, predict cognitive performance and anxiety in healthy humans [84, 85]. However, the connection between transcriptomic changes presented in this study and the energy metabolism still needs to be further consolidated.
The mitochondria-related gene network of naïve LBZ revealed that Ndufa10 is predicted to be associated with genes encoding OXPHOS subunits (Ndufa11, Cox6a1, and Atp5g1) and genes that regulate mitochondrial respiratory capacity, such as Cisd1 and Etfb. The Cisd1 gene, which encodes CDGSH iron–sulfur domain protein 1 in the mitochondrial outer membrane, regulates oxidative capacity and metabolism, possibly serving as a redox and pH sensor for mitochondrial function [86]. Notably, presynaptic mitochondrial proteome comparing PND7 and PND42 revealed increased Cisd1 levels at adolescence [87]. The electron-transfer flavoprotein b, a protein encoded by the Etfb gene, mediates the transfer of electrons from a series of mitochondrial dehydrogenases to the respiratory chain and is required for normal mitochondrial fatty acid oxidation and amino acid metabolism [88].
In stressed animals, our gene association network analysis revealed that the Cox6a1 gene is predicted to interact with other DEGs for complex I, III, and V subunits. Also, it is associated with the Chchd6 (Coiled-coil-helix-coiled-coil-helix domain-containing protein 6) gene, which is a component of the MICOS complex, a large protein complex of the mitochondrial inner membrane that maintains crista junctions and the formation of contact sites to the outer membrane [89]. Although no stress-induced psychiatry disorder has so far been associated with changes in Chchd6 gene expression, dysfunction of MICOS and OXPHOS impairment were found in neuronal cells lacking Disc1 (disrupted in Schizophrenia-1) gene [90]. Also, Cox6a1 was predicted to interact with a ribosomal protein gene (Rps3), which reduces cellular reactive oxygen species (ROS) levels and mitochondrial DNA damage when located in the mitochondrion. Furthermore, adolescent stress led to the down-regulation of Fis1 and Tomm20 genes, both key mediators of mitochondrial fission and mitochondrial translocase system, respectively. We also found a down-regulation of the Nlrx1 gene regulated in stressed animals compared to naïve LBZ. Nlrx1 is involved in oxidative stress and mitochondrial dynamics and detects mitochondrial protein-importing stress [91].
At the functional level, the behavioral variability among naïve animals and their mitochondrial gene signature was associated with reduced PFC OXPHOS capacity and changes in hydrogen peroxide production rate. Indeed, high-anxious outbred naïve rats exhibit reduced mitochondrial complex I and II proteins and respiratory capacity in the nucleus accumbens [30, 31]. Also, significant individual differences in mitochondrial content and respiratory chain activities across brain areas were described in inbred naïve mice not exposed to stressors [55].
Emerging evidence indicates that stress can impact mitochondrial function in different brain regions [39]. One day after stress (PND 41), the OXPHOS capacity of PFC mitochondria increased in stressed animals, accompanied by elevated hydrogen peroxide production. However, 10 days after stress exposure (PND 51), we found a decrease in OXPHOS capacity and reduced production of hydrogen peroxide linked to complex I activity. These findings suggest that adaptive changes in cortical energy metabolism, such as increased OXPHOS capacity after stress, may precede constitutive defects in mitochondrial function. Stress during critical neurodevelopmental periods, such as adolescence, can lead to a glutamatergic overdrive onto fast-spiking GABAergic interneurons expressing parvalbumin (PV), potentially changing the balance of excitation and inhibition (E/I) in cortical circuits that support cognitive processes [92,93,94]. PV interneurons are highly vulnerable to stress, particularly during neurodevelopment, and its reduction in cortical regions of animal models to study SCZ is accompanied by oxidative stress [95]. Although our adolescent stress protocol also negatively impacts PV-positive cells [24], this study lacks cell type specificity. In the future, it will also be essential to perform similar studies in female animals, especially under stressful conditions, as females are affected when the same stress protocol is applied later in adolescence [36].
Overall, our results provide evidence that distinct behavioral phenotypes in adolescent naïve animals and stressed animals are associated with diversity in mitochondrial features. While behavioral individualities, as characterized by LBZ vs. HBZ, may be related to inherited variations in mitochondrial gene expression and respiration, stress-associated changes are likely adaptative. Although further causal experiments are required, our study provides new insights into potential mechanisms of this variability, focusing on PFC mitochondrial-associated processes.
Data availability
The data that support the findings of this study and the R script analyses are available from the corresponding author upon reasonable request. Read counts of PFC RNA-sequencing are available on https://doi.org/10.6084/m9.figshare.24125793.v1.
References
Spear LP. Neurobehavioral changes in adolescence. Curr Dir Psychol Sci. 2000;9:111–4.
Bitzenhofer SH, Pöpplau JA, Chini M, Marquardt A, Hanganu-Opatz IL. A transient developmental increase in prefrontal activity alters network maturation and causes cognitive dysfunction in adult mice. Neuron. 2021;109:1350–1364.e6.
Van Kerkhof LWM, Damsteegt R, Trezza V, Voorn P, Vanderschuren LJMJ. Social play behavior in adolescent rats is mediated by functional activity in medial prefrontal cortex and striatum. Neuropsychopharmacology. 2013;38:1899–909.
Gee DG, Bath KG, Johnson CM, Meyer HC, Murty VP, van den Bos W, et al. Neurocognitive development of motivated behavior: dynamic changes across childhood and adolescence. J Neurosci. 2018;38:9433–45.
Sowell ER, Thompson PM, Holmes CJ, Jernigan TL, Toga AW. In vivo evidence for post-adolescent brain maturation in frontal and striatal regions. Nat Neurosci. 1999;2:859–61.
Arruda-Carvalho M, Wu WC, Cummings KA, Clem RL. Optogenetic examination of prefrontal-amygdala synaptic development. J Neurosci. 2017;37:2976–85.
Tamnes CK, Fjell AM, Westlye LT, Østby Y, Walhovd KB. Becoming consistent: developmental reductions in intraindividual variability in reaction time are related to white matter integrity. J Neurosci. 2012;32:972–82.
Luna B, Marek S, Larsen B, Tervo-Clemmens B, Chahal R. An integrative model of the maturation of cognitive control. Annu Rev Neurosci. 2015;38:151–70.
Guskjolen A, Josselyn SA, Frankland PW. Age-dependent changes in spatial memory retention and flexibility in mice. Neurobiol Learn Mem. 2017;143:59–66.
Larsen B, Luna B. Adolescence as a neurobiological critical period for the development of higher-order cognition. Neurosci Biobehav Rev. 2018;94:179–95.
Larrieu T, Cherix A, Lei H, Gruetter R. Hierarchical status predicts behavioral vulnerability and nucleus accumbens metabolic profile following report hierarchical status predicts behavioral vulnerability and nucleus accumbens metabolic profile following chronic social defeat stress. Curr Biol. 2017;27:2202–10.
Bijlsma A, Omrani A, Spoelder M, Verharen JPH, Bauer L, Cornelis C, et al. Social play behavior is critical for the development of prefrontal inhibitory synapses and cognitive flexibility in rats. J Neurosci. 2022;42:8716–28.
Bicks LK, Peng M, Taub A, Akbarian S, Morishita H. An adolescent sensitive period for social dominance hierarchy plasticity is regulated by cortical plasticity modulators in mice. Front Neural Circuits 2021;15. https://doi.org/10.3389/FNCIR.2021.676308.
Goldenberg D, Telzer EH, Lieberman MD, Fuligni AJ, Galván A. Greater response variability in adolescents is associated with increased white matter development. Soc Cogn Affect Neurosci. 2017;12:436–44.
Sturman DA, Moghaddam B. Reduced neuronal inhibition and coordination of adolescent prefrontal cortex during motivated behavior. J Neurosci. 2011;31:1471–8.
Caras ML, Sanes DH. Neural variability limits adolescent skill learning. J Neurosci. 2019;39:2889–902.
Toledo-Rodriguez M, Sandi C. Stress during adolescence increases novelty seeking and risk-taking behavior in male and female rats. Front Behav Neurosci. 2011;5:1–10.
Papilloud A, Guillot de Suduiraut I, Zanoletti O, Grosse J, Sandi C. Peripubertal stress increases play fighting at adolescence and modulates nucleus accumbens CB1 receptor expression and mitochondrial function in the amygdala. Transl Psychiatry 2018;8. https://doi.org/10.1038/s41398-018-0215-6.
Papilloud A, Veenit V, Tzanoulinou S, Riccio O, Zanoletti O, Guillot de Suduiraut I, et al. Peripubertal stress-induced heightened aggression: modulation of the glucocorticoid receptor in the central amygdala and normalization by mifepristone treatment. Neuropsychopharmacology. 2019;44:674–82.
Márquez C, Poirier GL, Cordero MI, Larsen MH, Groner A, Marquis J, et al. Peripuberty stress leads to abnormal aggression, altered amygdala and orbitofrontal reactivity and increased prefrontal MAOA gene expression. Transl Psychiatry 2013;3. https://doi.org/10.1038/tp.2012.144.
Walker SE, Sandi C. Long-term programing of psychopathology-like behaviors in male rats by peripubertal stress depends on individual’s glucocorticoid responsiveness to stress. Stress. 2018;21:433–42.
Veenit V, Cordero MI, Tzanoulinou S, Sandi C. Increased corticosterone in peripubertal rats leads to long-lasting alterations in social exploration and aggression. Front Behav Neurosci. 2013;7:1–10.
Veenit V, Riccio O, Sandi C. CRHR1 links peripuberty stress with deficits in social and stress-coping behaviors. J Psychiatr Res. 2014;53:1–7.
Gomes FV, Zhu X, Grace AA. The pathophysiological impact of stress on the dopamine system is dependent on the state of the critical period of vulnerability. Mol Psychiatry. 2019;25:3278–91.
Gomes FV, Grace AA. Adolescent stress as a driving factor for schizophrenia development—a basic science perspective. Schizophr Bull. 2017;43:486–9.
Gomes FV, Grace AA. Prefrontal cortex dysfunction increases susceptibility to schizophrenia-like changes induced by adolescent stress exposure. Schizophr Bull. 2017;43:592–600.
Harris LW, Lockstone HE, Khaitovich P, Weickert CS, Webster MJ, Bahn S. Gene expression in the prefrontal cortex during adolescence: implications for the onset of schizophrenia. BMC Med Genom. 2009;14:1–14.
Ene HM, Karry R, Farfara D, Ben-Shachar D. Mitochondria play an essential role in the trajectory of adolescent neurodevelopment and behavior in adulthood: evidence from a schizophrenia rat model. Mol Psychiatry 2022. https://doi.org/10.1038/s41380-022-01865-4.
Fame RM, Lehtinen MK. Mitochondria in early forebrain development: from neurulation to mid-corticogenesis. Front Cell Dev Biol. 2021;9:1–22.
Hollis F, Van Der Kooij MA, Zanoletti O, Lozano L, Cantó C, Sandi C. Mitochondrial function in the brain links anxiety with social subordination. Proc Natl Acad Sci USA. 2015;112:15486–91.
Gebara E, Zanoletti O, Ghosal S, Grosse J, Schneider BL, Knott G, et al. Mitofusin-2 in the nucleus accumbens regulates anxiety and depression-like behaviors through mitochondrial and neuronal actions. Biol Psychiatry. 2021;89:1033–44.
Ülgen DH, Ruigrok SR, Sandi C. Powering the social brain: mitochondria in social behaviour. Curr Opin Neurobiol. 2023;79:102675.
Filiou MD, Sandi C. Anxiety and brain mitochondria: a bidirectional crosstalk. Trends Neurosci. 2019;42:573–88.
Misiewicz Z, Iurato S, Kulesskaya N, Salminen L, Rodrigues L, Maccarrone G, et al. Multi-omics analysis identifies mitochondrial pathways associated with anxiety-related behavior. PLoS ONE. 2019;15:1–33.
Klinger K, Gomes FV, Rincón-Cortés M, Grace AA. Female rats are resistant to the long-lasting neurobehavioral changes induced by adolescent stress exposure. Eur Neuropsychopharmacol. 2019;29:1127–37.
Zhu X, Grace AA. Sex- and exposure age-dependent effects of adolescent stress on ventral tegmental area dopamine system and its afferent regulators. Mol Psychiatry. 2023;28:611.
Cavichioli AM, Santos-Silva T, Grace AA, Guimarães FS, Gomes FV. Levetiracetam attenuates adolescent stress-induced behavioral and electrophysiological changes associated with schizophrenia in adult rats. Schizophr Bull. 2022;49:68–77.
Guilloux JP, Seney M, Edgar N, Sibille E. Integrated behavioral z-scoring increases the sensitivity and reliability of behavioral phenotyping in mice: relevance to emotionality and sex. J Neurosci Methods. 2011;197:21–31.
Weger M, Alpern D, Cherix A, Ghosal S, Grosse J, Russeil J, et al. Mitochondrial gene signature in the prefrontal cortex for differential susceptibility to chronic stress. Sci Rep. 2020;10:1–15.
Alpern D, Gardeux V, Russeil J, Mangeat B, Meireles-Filho ACA, Breysse R, et al. BRB-seq: ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing. Genome Biol. 2019;20:1–15.
Wingett SW, Andrews S. Fastq screen: a tool for multi-genome mapping and quality control. F1000Res. 2018;7:1–13.
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–15.
Liao Y, Smyth GK, Shi W. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:1–17.
Anders S, Huber W. Differential expression and sequence-specific interaction of karyopherin α with nuclear localization sequences. Genome Biol. 2010;11:1–12.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B (Methodol). 1995;57:289–300.
Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216.
Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27.
Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. BioSystems. 2015;127:67–72.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models. Genome Res. 2003;13:2498–504.
Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–D646.
Binder JX, Pletscher-frankild S, Tsafou K, Stolte C, Donoghue IO, Schneider R, et al. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database. 2014;2014:1–9.
Larrieu T, Cherix A, Duque A, Rodrigues J, Lei H, Gruetter R, et al. Hierarchical status predicts behavioral vulnerability and nucleus accumbens metabolic profile following chronic social defeat stress. Curr Biol. 2017;27:2202–2210.e4.
Castro JE, Diessler S, Varea E, Márquez C, Larsen MH, Cordero MI, et al. Personality traits in rats predict vulnerability and resilience to developing stress-induced depression-like behaviors, HPA axis hyper-reactivity and brain changes in pERK1/2 activity. Psychoneuroendocrinology. 2012;37:1209–23.
Rosenberg AM, Saggar M, Monzel AS, Devine J, Rogu P, Limoges A, et al. Brain mitochondrial diversity and network organization predict anxiety-like behavior in male mice. Nat Commun. 2023;14:1–19.
Cowen L, Ideker T, Raphael BJ, Sharan R. Network propagation: a universal amplifier of genetic associations. Nat Rev Genet. 2017;18:551–62.
Tian W, Zhang LV, Taşan M, Gibbons FD, King OD, Park J, et al. Combining guilt-by-association and guilt-by-profiling to predict Saccharomyces cerevisiae gene function. Genome Biol 2008;9. https://doi.org/10.1186/GB-2008-9-S1-S7.
Bobba-Alves N, Juster RP, Picard M. The energetic cost of allostasis and allostatic load. Psychoneuroendocrinology. 2022;146:105951.
Picard M, Juster RP, McEwen BS. Mitochondrial allostatic load puts the ‘gluc’ back in glucocorticoids. Nat Rev Endocrinol. 2014;10:303–10.
Morató L, Astori S, Zalachoras I, Rodrigues J, Ghosal S, Huang W, et al. ENAMPT actions through nucleus accumbens NAD+/SIRT1 link increased adiposity with sociability deficits programmed by peripuberty stress. Sci Adv. 2022;8:1–19.
Poirier GL, Imamura N, Zanoletti O, Sandi C. Social deficits induced by peripubertal stress in rats are reversed by resveratrol. J Psychiatr Res. 2014;57:157–64.
Walker SE, Wood TC, Cash D, Mesquita M, Williams SCR, Sandi C. Alterations in brain microstructure in rats that develop abnormal aggression following peripubertal stress. Eur J Neurosci. 2018;48:1818–32.
Toledo-Rodriguez M, Pitiot A, Paus T, Sandi C. Stress during puberty boosts metabolic activation associated with fear-extinction learning in hippocampus, basal amygdala and cingulate cortex. Neurobiol Learn Mem. 2012;98:93–101.
Milner LC, Crabbe JC. Three murine anxiety models: results from multiple inbred strain comparisons. Genes Brain Behav. 2008;7:496–505.
van der Goot MH, Keijsper M, Baars A, Drost L, Hendriks J, Kirchhoff S, et al. Inter-individual variability in habituation of anxiety-related responses within three mouse inbred strains. Physiol Behav. 2021;239:1–16.
Zalachoras I, Astori S, Meijer M, Grosse J, Zanoletti O, De Suduiraut IG, et al. Opposite effects of stress on effortful motivation in high and low anxiety are mediated by CRHR1 in the VTA. Sci Adv 2022;8: SPOTLIGHTS.
Herrero AI, Sandi C, Venero C. Individual differences in anxiety trait are related to spatial learning abilities and hippocampal expression of mineralocorticoid receptors. Neurobiol Learn Mem. 2006;86:150–9.
Sachser N, Hennessy MB, Kaiser S. The adaptive shaping of social behavioural phenotypes during adolescence. Biol Lett. 2018;14:1–7.
Jones CE, Monfils MH. Dominance status predicts social fear transmission in laboratory rats. Anim Cogn. 2016;19:1051–69.
Van Den Berg CL, Hol T, Van Ree JM, Spruijt BM, Everts H, Koolhaas JM. Play is indispensable for an adequate development of coping with social challenges in the rat. Dev Psychobiol. 1999;34:129–38.
Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, et al. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302.
Picard M, McEwen BS, Epel ES, Sandi C. An energetic view of stress: focus on mitochondria. Front Neuroendocrinol. 2018;49:72–85.
Hjelm BE, Rollins B, Mamdani F, Lauterborn JC, Kirov G, Lynch G, et al. Evidence of mitochondrial dysfunction within the complex genetic etiology of schizophrenia. Complex Psychiatry. 2015;1:201–19.
Haghighatfard A, Andalib S, Amini Faskhodi M, Sadeghi S, Ghaderi AH, Moradkhani S, et al. Gene expression study of mitochondrial complex I in schizophrenia and paranoid personality disorder. World J Biol Psychiatry. 2018;19:S133–S146.
Anitha A, Nakamura K, Thanseem I, Matsuzaki H, Miyachi T, Tsujii M, et al. Downregulation of the expression of mitochondrial electron transport complex genes in autism brains. Brain Pathol. 2013;23:294–302.
Akarsu S, Torun D, Bolu A, Erdem M, Kozan S, Ak M, et al. Mitochondrial complex I and III gene mRNA levels in schizophrenia, and their relationship with clinical features. J Mol Psychiatry. 2014;2:2–7.
Davis LK, Gamazon ER, Kistner-Griffin E, Badner JA, Liu C, Cook EH, et al. Loci nominally associated with autism from genome-wide analysis show enrichment of brain expression quantitative trait loci but not lymphoblastoid cell line expression quantitative trait loci. Mol Autism. 2012;3:1–11.
Sanz‐Morello B, Pfisterer U, Winther Hansen N, Demharter S, Thakur A, Fujii K, et al. Complex IV subunit isoform COX 6A2 protects fast‐spiking interneurons from oxidative stress and supports their function. EMBO J. 2020;39:1–21.
Yang S, Wu P, Xiao J, Jiang L. Overexpression of COX6B1 protects against I/R-induced neuronal injury in rat hippocampal neurons. Mol Med Rep. 2019;19:4852–62.
Khadimallah I, Jenni R, Cabungcal JH, Cleusix M, Fournier M, Beard E, et al. Mitochondrial, exosomal miR137-COX6A2 and gamma synchrony as biomarkers of parvalbumin interneurons, psychopathology, and neurocognition in schizophrenia. Mol Psychiatry. 2022;27:1192–204.
Puertas-Frías G, del Arco A, Pardo B, Satrústegui J, Contreras L. Mitochondrial movement in Aralar/Slc25a12/AGC1 deficient cortical neurons. Neurochem Int. 2019;131:1–15.
Kanellopoulos AK, Mariano V, Spinazzi M, Woo YJ, McLean C, Pech U, et al. Aralar sequesters GABA into hyperactive mitochondria, causing social behavior deficits. Cell. 2020;180:1178–1197.e20.
Andersen JV, Markussen KH, Jakobsen E, Schousboe A, Waagepetersen HS, Rosenberg PA, et al. Glutamate metabolism and recycling at the excitatory synapse in health and neurodegeneration. Neuropharmacology. 2021;196:108719.
Strasser A, Luksys G, Xin L, Pessiglione M, Gruetter R, Sandi C. Glutamine-to-glutamate ratio in the nucleus accumbens predicts effort-based motivated performance in humans. Neuropsychopharmacology. 2020;45:2048–57.
Strasser A, Xin L, Gruetter R, Sandi C. Nucleus accumbens neurochemistry in human anxiety: a 7 T 1H-MRS study. Eur Neuropsychopharmacol. 2019;29:365–75.
Geldenhuys WJ, Long TE, Saralkar P, Iwasaki T, Nuñez RAA, Nair RR, et al. Crystal structure of the mitochondrial protein mitoNEET bound to a benze-sulfonide ligand. Commun Chem. 2019;2:1–9.
Stauch KL, Villeneuve LM, Totusek S, Lamberty B, Ciborowski P, Fox HS. Quantitative proteomics of presynaptic mitochondria reveal an overexpression and biological relevance of neuronal MitoNEET in postnatal brain development. Dev Neurobiol. 2019;79:370–86.
Henriques BJ, Katrine Jentoft Olsen R, Gomes CM, Bross P. Electron transfer flavoprotein and its role in mitochondrial energy metabolism in health and disease. Gene. 2021;776:145407.
Anand R, Reichert AS, Kondadi AK. Emerging roles of the MICOS complex in cristae dynamics and biogenesis. Biology (Basel) 2021;10. https://doi.org/10.3390/BIOLOGY10070600.
Piñero-Martos E, Ortega-Vila B, Pol-Fuster J, Cisneros-Barroso E, Ruiz-Guerra L, Medina-Dols A, et al. Disrupted in schizophrenia 1 (DISC1) is a constituent of the mammalian mitochondrial contact site and cristae organizing system (MICOS) complex, and is essential for oxidative phosphorylation. Hum Mol Genet. 2016;25:4157.
Stokman G, Kors L, Bakker PJ, Rampanelli E, Claessen N, Teske GJD, et al. NLRX1 dampens oxidative stress and apoptosis in tissue injury via control of mitochondrial activity. J Exp Med. 2017;214:2405–20.
Tzanoulinou S, García-Mompó C, Riccio O, Grosse J, Zanoletti O, Dedousis P, et al. Neuroligin-2 expression in the prefrontal cortex is involved in attention deficits induced by peripubertal stress. Neuropsychopharmacology. 2016;41:751–61.
Tzanoulinou S, Riccio O, de Boer MW, Sandi C. Peripubertal stress-induced behavioral changes are associated with altered expression of genes involved in excitation and inhibition in the amygdala. Transl Psychiatry. 2014;4:1–9.
Tzanoulinou S, García-Mompó C, Castillo-Gómez E, Veenit V, Nacher J, Sandi C. Long-term behavioral programming induced by peripuberty stress in rats is accompanied by gabaergic-related alterations in the amygdala. PLoS ONE 2014;9. https://doi.org/10.1371/journal.pone.0094666.
Steullet P, Cabungcal JH, Coyle J, Didriksen M, Gill K, Grace AA, et al. Oxidative stress-driven parvalbumin interneuron impairment as a common mechanism in models of schizophrenia. Mol Psychiatry. 2017;22:936–43.
Acknowledgements
The authors thank Marco Antonio de Carvalho, Eleni Tamburus Gomes, and Eliane Aparecida Antunes Maciel for technical assistance. The authors acknowledge Sabrina Baroni’s technical support in performing the RNA integrity analysis.
Funding
This work received financial support from the São Paulo Research Foundation (FAPESP-2018/17597-3 to FVG), the International Brain Research Organization (IBRO Return Home Fellowship to FVG), and Coordination for the Improvement of Higher Education Personnel—Brazil (CAPES)—Finance Code 001. TSS acknowledges fellowships from CAPES (88887.334572/2019-00) and its Institutional Internationalization Program—CAPES-Print, Brazil (88887.575008/2020-00). CS received grants from the Swiss National Science Foundation (SNSF) (176206 and 1979042; NCCR Synapsys: 51NF40-185897) and ERA-NET (Biostress; SNSF no. 31NE30_189061) and intramural funding from the EPFL. CFBL acknowledges a fellowship from CAPES (88887.699520/2022-00). LCA acknowledges FAPESP support (grant 2018/10089-2) and the National Council for Scientific and Technological Development (CNPq). FSG acknowledges FAPESP support (grant 2017/24304-0).
Author information
Authors and Affiliations
Contributions
TSS intellectually conceptualized the idea of the study, interpreted critically all the results to write a first draft of the manuscript; revised the manuscript critically; participated actively in all the conception and design of the study; actively performed the rodent model, bioinformatic and biomolecular experiments; performed all analyses; created the figures and tables. DHU conceptualized and designed the transcriptomic experiment; contributed to managing the RNA-seq experiment; performed the bioinformatic analysis; interpreted bioinformatic data and revised the manuscript. CFBL conceptualized and designed the experiments for mitochondrial function and redox dysregulation; performed and critically interpreted all the mitochondrial respiratory capacity measurements (high-resolution respirometry), Amplex™ Red, and MitoSOX™ assays; performed the RNA isolation; critically revised the manuscript. LCA contributed to managing the high-resolution respirometry; interpreted this data critically, and revised the manuscript. FSG contributed to managing animal and biomolecular resources and critically revised the manuscript. CS conceptualized the behavioral and bioinformatic analyses and critically revised the manuscript. FVG intellectually conceptualized the idea of the study and critique.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Santos-Silva, T., Hazar Ülgen, D., Lopes, C.F.B. et al. Transcriptomic analysis reveals mitochondrial pathways associated with distinct adolescent behavioral phenotypes and stress response. Transl Psychiatry 13, 351 (2023). https://doi.org/10.1038/s41398-023-02648-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-023-02648-3