Abstract
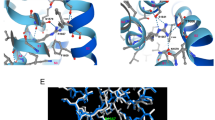
Butyrylcholinesterase (BChE) deficiency is characterized by prolonged apnea after the use of muscle relaxants (suxamethonium or mivacurium) in patients who have mutations in the BCHE gene. Here, we report the characterization of four BCHE mutations associated with prolonged effect of suxamethonium (amino acid numbering based on the matured enzyme): p.20delValPheGlyGlyThrValThr, p.Leu88His, p.Ile140del and p.Arg386Cys. Expression of recombinant BCHE mutants, kinetic analysis and molecular dynamics were undertaken to understand how these mutations induce BChE deficiency. Three of the mutations studied (p.20delValPheGlyGlyThrValThr, p.Ile140del and p.Arg386Cys) lead to a “silent” BChE phenotype. Recombinant BCHE expression studies for these mutants revealed BChE activity levels comparable to untransfected cells. Only the last one (hBChE-L88H) presented BChE activity in the transfected cell culture medium. This BChE mutant (p.Leu88His) is associated with a lower kcat value compare to the wild-type enzyme. Molecular dynamics simulations analyses suggest that a destabilization of a structure implicated in enzyme activity (Ω-loop) can explain the modification of the kinetic parameter of the mutated protein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lockridge O. Review of human butyrylcholinesterase structure, function,genetic variants, history of use in the clinic, and potential therapeutic uses. Pharmacol Ther. 2015;148:34–46.
Schopfer LM, Lockridge O, Brimijoin S. Pure human butyrylcholinesterase hydrolyzes octanoyl ghrelin to desacyl ghrelin. Gen Comp Endocrinol. 2015;224:61–8.
Chen VP, Gao Y, Geng L, Parks RJ, Pang YP, Brimijoin S. Plasma butyrylcholinesterase regulates ghrelin to control aggression. Proc Natl Acad Sci USA. 2015;112:2251–6.
Brimijoin S, Chen VP, Pang YP, Geng L, Gao Y. Physiological roles for butyrylcholinesterase: a BChE-ghrelin axis. Chem Biol Interact. 2016;259:271–5.
Forbat A, Lehmann H, Silk E. Prolonged apnoea following injection of succinylcholine. Lancet. 1953;265:1067–8.
Kalow W, Genest K. A method for the detection of atypical forms of human serum cholinesterase; determination of dicucaine number. Can J Biochem Physiol. 1957;35:339–46.
Delacour H, Ceppa F. Déficit en butyrylcholinestérase: quelle démarche diagnostique, quelles conséquences pour le patient? Anest Reanim. 2017;3:99–100.
Masson P, Legrand P, Bartels CF, Froment MT, Schopfer LM, Lockridge O. Role of aspartate 70 and tryptophan 82 in binding of succinyldithiocholine to human butyrylcholinesterase. Biochemistry. 1997;36:2266–77.
Podoly E, Shalev DE, Shenhar-Tsarfaty S, Bennett ER, Ben Assayag E, Wilgus H, et al. The butyrylcholinesterase K variant confers structurally derived risks for Alzheimer pathology. J Biol Chem. 2009;284:17170–9.
David SM, Venkatesan SK, Boopathy R. An Indian butyrylcholinesterase variant L307P is not structurally stable: a molecular dynamics simulation study. Chem Biol Interact. 2013;203:30–5.
Delacour H, Lushchekina S, Mabboux I, Ceppa F, Masson P, Schopfer LM, et al. Characterization of a novel butyrylcholinesterase point mutation (p.Ala34Val), “silent” with mivacurium. Biochem Pharmacol. 2014;92:476–83.
Delacour H, Lushchekina S, Mabboux I, Bousquet A, Ceppa F, Schopfer LM, et al. Characterization of a novel BCHE “silent” allele: pointmutation (p.Val204Asp) causes loss of activity and prolonged apnea with suxamethonium. PLoS ONE. 2014;9:e101552.
Lushchekina S, Delacour H, Lockridge O, Masson P. Human butyrylcholinesterase polymorphism: Molecular modeling. Int J Risk Saf Med. 2015;27:S80–1.
Radic Z, Pickering NA, Vellom DC, Camp S, Taylor P. Three distinct domains in the cholinesterase molecule confer selectivity for acetyl- and butyrylcholinesterase inhibitors. Biochemistry. 1993;32:12074–84.
Nachon F, Nicolet Y, Viguie N, Masson P, Fontecilla-Camps JC, Lockridge O. Engineering of a monomeric and low-glycosylated form of human butyrylcholinesterase: expression, purification, characterization and crystallization. Eur J Biochem. 2002;269:630–7.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–47.
Karnovsky MJ, Roots LA. “Direct-coloring” thiocholine method for cholinesterases. J Histochem Cytochem. 1964;12:219–21.
Chiu J, March PE, Lee R, Tillett D. Site-directed, Ligase-Independent Mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 2004;32:e174.
Lockridge O, Blong RM, Masson P, Froment MT, Millard CB, Broomfield CA. A single amino acid substitution, Gly117His, confers phosphotriesterase (organophosphorus acid anhydride hydrolase) activity on human butyrylcholinesterase. Biochemistry. 1997;36:786–95.
Brazzolotto X, Wandhammer M, Ronco C, Trovaslet M, Jean L, Lockridge O, et al. Human butyrylcholinesterase produced in insect cells: huprine-based affinity purification and crystal structure. FEBS J. 2012;279:2905–16.
Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2014;1137:1–15.
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25.
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–25.
Nicolet Y, Lockridge O, Masson P, Fontecilla-Camps JC, Nachon F. Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products. J Biol Chem. 2003;278:41141–7.
Leung MR, van Bezouwen LS, Schopfer LM, Sussman JL, Silman I, Lockridge O, et al. Cryo-EM structure of the native butyrylcholinesterase tetramer reveals a dimer of dimers stabilized by a superhelical assembly. Proc Natl Acad Sci USA. 2018;115:13270–5.
Masson P, Carletti E, Nachon F. Structure, activities and biomedical applications of human butyrylcholinesterase. Protein Pept Lett. 2009;16:1215–24.
Yen T, Nightingale BN, Burns JC, Sullivan DR, Stewart PM. Butyrylcholinesterase (BCHE) genotyping for post-succinylcholine apnea in an Australian population. Clin Chem. 2003;49:1297–308.
Karczewski K, Francioli L, Tiao G, Cummings B. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. 2019. https://www.biorxiv.org/content/10.1101/531210v4.
Masson P, Froment MT, Bartels CF, Lockridge O. Asp70 in the peripheral anionic site of human butyrylcholinesterase. Eur J Biochem. 1996;235:36–48.
De Jaco A, Comoletti D, Kovarik Z, Gaietta G, Radic Z, Lockridge O, et al. A mutation linked with autism reveals a common mechanism of endoplasmic reticulum retention for the alpha,beta-hydrolase fold protein family. J Biol Chem. 2006;281:9667–76.
Zhang X, Lin H, Zhao H, Hao Y, Mort M, Cooper DN, et al. Impact of human pathogenic micro-insertions and micro-deletions on post-transcriptional regulation. Hum Mol Genet. 2014;23:3024–34.
The Uniprot Consortium. Uniprot: a worldwild hub of protein knowledge. Nucleic Acid Res. 2019;47:D506–15.
Mikami LR, Wieseler S, Souza RL, Schopfer LM, Lockridge O, Chautard-Freire-Maia EA. Expression of three naturally occurring genetic variants (G75R, E90D, I99M) of the BCHE gene of human butyrylcholinesterase. Pharmacogenet Genom. 2007;17:681–5.
Schrodinger LLC. The PyMOL molecular graphics system. Version. 2010;1:3r1.
Acknowledgements
XB, VG, AI and FN were supported by the French Ministry of Armed Forces (Direction Générale de l’Armement et Service de Santé des Armées) under project NBC-5-C-4210 and PDH-2-NRBC-3-C-3201.The authors would like to thank Prof. Oksana Lockrige and Dr. Eric Krejci for their technical advices.
Author information
Authors and Affiliations
Contributions
Concept and design: XB, HD. Data collection: SC, CS, VG, AI. Data analysis: XB, FN, HD. Writing: XB, HD, JK. Critical review: FN, FC.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Brazzolotto, X., Courcelle, S., Sauvanet, C. et al. Characterization of four BCHE mutations associated with prolonged effect of suxamethonium. Pharmacogenomics J 21, 165–173 (2021). https://doi.org/10.1038/s41397-020-00192-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-020-00192-7