Abstract
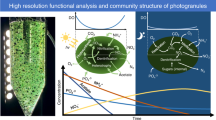
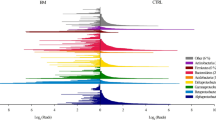
Granular biofilms producing medium-chain carboxylic acids (MCCA) from carbohydrate-rich industrial feedstocks harbor highly streamlined communities converting sugars to MCCA either directly or via lactic acid as intermediate. We investigated the spatial organization and growth activity patterns of MCCA producing granular biofilms grown on an industrial side stream to test (i) whether key functional guilds (lactic acid producing Olsenella and MCCA producing Oscillospiraceae) stratified in the biofilm based on substrate usage, and (ii) whether spatial patterns of growth activity shaped the unique, lenticular morphology of these biofilms. First, three novel isolates (one Olsenella and two Oscillospiraceae species) representing over half of the granular biofilm community were obtained and used to develop FISH probes, revealing that key functional guilds were not stratified. Instead, the outer 150–500 µm of the granular biofilm consisted of a well-mixed community of Olsenella and Oscillospiraceae, while deeper layers were made up of other bacteria with lower activities. Second, nanoSIMS analysis of 15N incorporation in biofilms grown in normal and lactic acid amended conditions suggested Oscillospiraceae switched from sugars to lactic acid as substrate. This suggests competitive-cooperative interactions may govern the spatial organization of these biofilms, and suggests that optimizing biofilm size may be a suitable process engineering strategy. Third, growth activities were similar in the polar and equatorial biofilm peripheries, leaving the mechanism behind the lenticular biofilm morphology unexplained. Physical processes (e.g., shear hydrodynamics, biofilm life cycles) may have contributed to lenticular biofilm development. Together, this study develops an ecological framework of MCCA-producing granular biofilms that informs bioprocess development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
16S rRNA gene sequences have been deposited to NCBI GenBank under accession numbers OP799682–OP799726.
References
Agler MT, Spirito CM, Usack JG, Werner JJ, Angenent LT. Chain elongation with reactor microbiomes: upgrading dilute ethanol to medium-chain carboxylates. Energy Environ Sci. 2012;5:8189–92.
Angenent LT, Richter H, Buckel W, Spirito CM, Steinbusch KJJ, Plugge CM, et al. Chain elongation with reactor microbiomes: open-culture biotechnology to produce biochemicals. Environ Sci Technol. 2016;50:2796–810.
Urban C, Xu J, Sträuber H, dos Santos Dantas TR, Mühlenberg J, Härtig C, et al. Production of drop-in fuels from biomass at high selectivity by combined microbial and electrochemical conversion. Energy Environ Sci. 2017;10:2231–44.
Roghair M, Strik DPBTB, Steinbusch KJJ, Weusthuis RA, Bruins ME, Buisman CJN. Granular sludge formation and characterization in a chain elongation process. Process Biochem. 2016;51:1594–8.
Carvajal-Arroyo JM, Candry P, Andersen SJ, Props R, Seviour T, Ganigué R, et al. Granular fermentation enables high-rate caproic acid production from solid-free thin stillage. Green Chem. 2019;21:1330–9.
Wu Q, Feng X, Chen Y, Liu M, Bao X. Continuous medium chain carboxylic acids production from excess sludge by granular chain-elongation process. J Hazard Mater. 2021;402:123471.
Mariën Q, Candry P, Hendriks E, Carvajal-Arroyo JM, Ganigué R. Substrate loading and nutrient composition steer caproic acid production and biofilm aggregation in high-rate granular reactors. J Environ Chem Eng. 2022;10:107727.
Mariën Q, Ulčar B, Verleyen J, Vanthuyne B, Ganigué R. High-rate conversion of lactic acid-rich streams to caproic acid in a fermentative granular system. Bioresour Technol. 2022;355:127250.
Smith AL, Stadler LB, Cao L, Love NG, Raskin L, Skerlos SJ. Navigating wastewater energy recovery strategies: a life cycle comparison of anaerobic membrane bioreactor and conventional treatment systems with anaerobic digestion. Environ Sci Technol. 2014;48:5972–81.
Dewhirst FE, Paster BJ, Tzellas N, Coleman B, Downes J, Spratt DA, et al. Characterization of novel human oral isolates and cloned 16S rDNA sequences that fall in the family Coriobacteriaceae: description of Olsenella gen. nov., reclassification of Lactobacillus uli as Olsenella uli comb. nov. and description of Olsenella. Int J Syst Evol Microbiol. 2001;51:1797–804.
Zhu X, Zhou Y, Wang Y, Wu T, Li X, Li D, et al. Production of high-concentration n-caproic acid from lactate through fermentation using a newly isolated Ruminococcaceae bacterium CPB6. Biotechnol Biofuels. 2017;10:1–12.
Kim BC, Jeon BS, Kim S, Kim H, Um Y, Sang BI. Caproiciproducens galactitolivorans gen. nov., sp. nov., a bacterium capable of producing caproic acid from galactitol, isolated from a wastewater treatment plant. Int J Syst Evol Microbiol. 2015;65:4902–8.
Kang S, Kim H, Jeon BS, Choi O, Sang BI. Chain elongation process for caproate production using lactate as electron donor in megasphaera hexanoica. Bioresour Technol. 2022;346:126660.
Wang H, Li X, Wang Y, Tao Y, Lu S, Zhu X, et al. Improvement of n-caproic acid production with Ruminococcaceae bacterium CPB6: Selection of electron acceptors and carbon sources and optimization of the culture medium. Micro Cell Fact. 2018;17:1–9.
Candry P, Ganigué R. Chain elongators, friends, and foes. Curr Opin Biotechnol. 2021;67:99–110.
Royce LA, Liu P, Stebbins MJ, Hanson BC, Jarboe LR. The damaging effects of short-chain fatty acids on Escherichia coli membranes. Appl Microbiol Biotechnol. 2013;97:8317–27.
Candry P, van Daele T, Denis K, Amerlinck Y, Andersen SJ, Ganigué R. et al. A novel high-throughput method for kinetic characterization of anaerobic bioproduction strains, applied to Clostridium kluyveri. Sci Rep. 2018;8:9724.
Winkler MKH, Kleerebezem R, Van Loosdrecht MCM. Integration of anammox into the aerobic granular sludge process for main stream wastewater treatment at ambient temperatures. Water Res. 2012;46:136–44.
Winkler MKH, Kleerebezem R, Strous M, Chandran K, Van Loosdrecht MCM. Factors influencing the density of aerobic granular sludge. Appl Microbiol Biotechnol. 2013;97:7459–68.
Zheng D, Angenent LT, Raskin L. Monitoring granule formation in anaerobic up-flow bioreactors using oligonucleotide hybridization probes. Biotechnol Bioeng. 2006;94:458–72.
Mcglynn SE, Chadwick GL, Kempes CP, Orphan VJ. Single-cell activity reveals direct electron transfer in methanotrophic consortia. Nature. 2015;526:531–5.
Dawson KS, Scheller S, Dillon JG, Orphan VJ. Stable isotope phenotyping via cluster analysis of NanoSIMS data as a method for characterizing distinct microbial ecophysiologies and sulfur-cycling in the environment. Front Microbiol. 2016;7:774.
Jeon BS, Kim BC, Um Y, Sang BI. Production of hexanoic acid from d-galactitol by a newly isolated Clostridium sp. BS-1. Appl Microbiol Biotechnol. 2010;88:1161–7.
Kopf SH, McGlynn SE, Green-Saxena A, Guan Y, Newman DK, Orphan VJ. Heavy water and 15N labelling with NanoSIMS analysis reveals growth rate-dependent metabolic heterogeneity in chemostats. Environ Microbiol. 2015;17:2542–56.
Fuchs BM, Pernthaler J, Amann R Single cell identification by fluorescencein situ hybridization. In: Reddy CA, Beveridgje TJ, Breznak JA, Marzluf GA, Schmidt TM, Snyder LR, editors. Methods for general and molecular microbiology. 3rd ed. Washington D.C. ASM Press; 2007. p. 886–96.
Polerecky L, Adam B, Milucka J, Musat N, Vagner T, Kuypers MMM. Look@NanoSIMS—a tool for the analysis of nanoSIMS data in environmental microbiology. Environ Microbiol. 2012;14:1009–23.
Andersen SJ, Candry P, Basadre T, Khor WC, Roume H, Hernandez-Sanabria E, et al. Electrolytic extraction drives volatile fatty acid chain elongation through lactic acid and replaces chemical pH control in thin stillage fermentation. Biotechnol Biofuels. 2015;8:1–14.
Gildemyn S, Verbeeck K, Slabbinck R, Andersen SJ, Prévoteau A, Rabaey K. Integrated production, extraction, and concentration of acetic acid from CO2 through microbial electrosynthesis. Environ Sci Technol Lett. 2015;2:325–8.
Geirnaert A, Wang J, Tinck M, Steyaert A, Van den Abbeele P, Eeckhaut V. et al. Interindividual differences in response to treatment with butyrate-producing Butyricicoccus pullicaecorum 25-3T studied in an in vitro gut model. FEMS Microbiol Ecol. 2015;91:fiv054
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–9.
Heuer H, Krsek M, Baker P, Smalla K, Wellington EMH. Analysis of actinomycete communities by specific amplification of genes encoding 16S rDNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–41.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9.
Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–26.
Rohwer RR, Hamilton JJ, Newton RJ, McMahon KD. TaxAss: leveraging a custom freshwater database achieves fine-scale taxonomic resolution. mSphere. 2018;3:10–128.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar A, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6.
Gu Y, Zhu X, Lin F, Shen C, Li Y, Ao L, et al. Caproicibacterium amylolyticum gen. Nov., sp. nov., a novel member of the family oscillospiraceae isolated from pit clay used for making Chinese strong aroma-type liquor. Int J Syst Evol Microbiol. 2021;71:004789.
Esquivel-Elizondo S, Bağcı C, Temovska M, Jeon BS, Bessarab I, Williams RBH. et al. The isolate caproiciproducens sp. 7D4C2 produces n-caproate at mildly acidic conditions from hexoses: genome and rbox comparison with related strains and chain-elongating bacteria. Front Microbiol. 2021;11:594524.
Flaiz M, Baur T, Brahner S, Poehlein A, Daniel R, Bengelsdorf FR. Caproicibacter fermentans gen. Nov., sp. nov., a new caproate-producing bacterium and emended description of the genus caproiciproducens. Int J Syst Evol Microbiol. 2020;70:4269–79.
Staats N, Stal LJ, Mur LR. Exopolysaccharide production by the epipelic diatom Cylindrotheca closterium: effects of nutrient conditions. J Exp Mar Biol Ecol. 2000;249:13–27.
Trego AC, Galvin E, Sweeney C, Dunning S, Murphy C, Mills S. et al. Growth and break-up of methanogenic granules suggests mechanisms for biofilm and community development. Front Microbiol. 2020;11:1126.
Leventhal GE, Boix C, Kuechler U, Enke TN, Sliwerska E, Holliger C. et al. Strain-level diversity drives alternative community types in millimetre-scale granular biofilms. Nat Microbiol. 2018;3:1295–303.
Allaart MT, Stouten GR, Sousa DZ, Kleerebezem R. Product inhibition and ph affect stoichiometry and kinetics of chain elongating microbial communities in sequencing batch bioreactors. Front Bioeng Biotechnol. 2021;9:693030.
Andersen SJ, Groof VD, Khor WC, Roume H, Props R, Coma M, et al. A clostridium Group IV species dominates and suppresses a mixed culture fermentation by tolerance to medium chain fatty acids products. Front Bioeng Biotechnol. 2017;5:1–10.
Scarborough MJ, Lawson CE, Hamilton JJ, Donohue TJ, Noguera DR. Metatranscriptomic and thermodynamic insights into medium-chain fatty acid production using an anaerobic microbiome. mSystems 2018;3:1–21.
Scarborough MJ, Hamilton JJ, Erb EA, Donohue TJ, Noguera DR. Diagnosing and predicting mixed-culture fermentations with unicellular and guild-based metabolic models. mSystems 2020;5:e00755–20.
Nguyen Quoc B, Armenta M, Carter JA, Bucher R, Sukapanpotharam P, Bryson SJ, et al. An investigation into the optimal granular sludge size for simultaneous nitrogen and phosphate removal. Water Res. 2021;198:117119.
Nguyen Quoc B, Wei S, Armenta M, Bucher R, Sukapanpotharam P, Stahl DA, et al. Aerobic granular sludge: impact of size distribution on nitrification capacity. Water Res. 2021;188:116445.
Krasňan V, Stloukal R, Rosenberg M, Rebroš M. Immobilization of cells and enzymes to LentiKats®. Appl Microbiol Biotechnol. 2016;100:2535–53.
Winkler MKH, Bassin JP, Kleerebezem R, de Bruin LMM, van den Brand TPH, van Loosdrecht MCM. Selective sludge removal in a segregated aerobic granular biomass system as a strategy to control PAO–GAO competition at high temperatures. Water Res. 2011;45:3291–9.
Winkler MKH, Kleerebezem R, Kuenen JG, Yang J, van Loosdrecht MCM. Segregation of biomass in cyclic anaerobic/aerobic granular sludge allows the enrichment of anaerobic ammonium oxidizing bacteria at low temperatures. Environ Sci Technol. 2011;45:7330–7.
Tay JH, Liu QS, Liu Y. The effect of upflow air velocity on the structure of aerobic granules cultivated in a sequencing batch reactor. Water Sci Technol. 2004;49:35–40.
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Deverux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–25.
Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe eub338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–44.
Acknowledgements
PC was supported by UGent Special Research Fund (BOF15/DOC/286) and the U.S. Department of Energy, Office of Science, Office of Biological & Environmental Research (#DE-SC0020356). RG is supported by the Special Research Fund of Ghent University (grant number BOF19/STA/044). GLC and VJO were supported by grants from the Department of Energy, Office of Science, Office of Biological and Environmental Research (#DE-SC0020373). VJO was also supported by the Life Sciences-Simons Collaboration on Principles of Microbial Ecosystems (PriME) Program from the Simons Foundation (Award 542393).
Author information
Authors and Affiliations
Contributions
Conceptualization: PC, GLC, and JMCA. Methodology: PC and GLC. Investigation: PC, GLC, and TL. Formal Analysis: PC and GLC. Supervision: VJO and KR. Writing—Original Draft: PC. Writing—Review & Editing: All authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Candry, P., Chadwick, G.L., Caravajal-Arroyo, J.M. et al. Trophic interactions shape the spatial organization of medium-chain carboxylic acid producing granular biofilm communities. ISME J 17, 2014–2022 (2023). https://doi.org/10.1038/s41396-023-01508-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-023-01508-8