Abstract
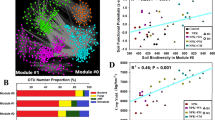
The microbiome function responses to land use change are important for the long-term prediction and management of soil ecological functions under human influence. However, it has remains uncertain how the biogeographic patterns of soil functional composition change when transitioning from natural steppe soils (NS) to agricultural soils (AS). We collected soil samples from adjacent pairs of AS and NS across 900 km of Mollisol areas in northeast China, and the soil functional composition was characterized using shotgun sequencing. AS had higher functional alpha-diversity indices with respect to KO trait richness and a higher Shannon index than NS. The distance-decay slopes of functional gene composition were steeper in AS than in NS along both spatial and environmental gradients. Land-use conversion from steppe to farmland diversified functional gene profiles both locally and spatially; it increased the abundances of functional genes related to labile carbon, but decreased those related to recalcitrant substrate mobilization (e.g., lignin), P cycling, and S cycling. The composition of gene functional traits was strongly driven by stochastic processes, while the degree of stochasticity was higher in NS than in AS, as revealed by the neutral community model and normalized stochasticity ratio analysis. Alpha-diversity of core functional genes was strongly related to multi-nutrient cycling in AS, suggesting a key relationship to soil fertility. The results of this study challenge the paradigm that the conversion of natural to agricultural habitat will homogenize soil properties and biology while reducing local and regional gene functional diversity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The raw sequence data of this study have been deposited into the Genome Sequence Archive (GSA) database under accession number of CRA004163, which were publicly accessible at. All code used for processing and analyzing the data is available in this GitHub repository [https://github.com/Microbion/2022-Conversion-of-steppe-to-agriculture].
References
Forrest JR, Thorp RW, Kremen C, Williams NM. Contrasting patterns in species and functional-trait diversity of bees in an agricultural landscape. J Appl Ecol. 2015;52:706–15.
Mbuthia LW, Acosta-Martínez V, DeBruyn J, Schaeffer S, Tyler D, Odoi E, et al. Long term tillage, cover crop, and fertilization effects on microbial community structure, activity: implications for soil quality. Soil Biol Biochem. 2015;89:24–34.
Xun W, Xu Z, Li W, Ren Y, Huang T, Ran W, et al. Long-term organic-inorganic fertilization ensures great soil productivity and bacterial diversity after natural to agricultural ecosystem conversion. J Microbiol. 2016;54:611–7.
Cornell CR, Zhang Y, Ning D, Wu L, Wagle P, Steiner JL, et al. Temporal dynamics of bacterial communities along a gradient of disturbance in a US Southern Plains agroecosystem. mBio 2022;13:e03829–21.
Borrelli P, Robinson DA, Panagos P, Lugato E, Yang JE, Alewell C, et al. Land use and climate change impacts on global soil erosion by water (2015-2070). Proc Natl Acad Sci USA. 2020;117:21994–2001.
Gupta A, Singh UB, Sahu PK, Paul S, Kumar A, Malviya D, et al. Linking soil microbial diversity to modern agriculture practices: a review. Int J Environ Res Public Health. 2022;19:3141.
Göpel J, Schüngel J, Stuch B, Schaldach R. Assessing the effects of agricultural intensification on natural habitats and biodiversity in Southern Amazonia. PloS One. 2020;15:e0225914.
Šálek M, Kalinová K, Daňková R, Grill S, Żmihorski M. Reduced diversity of farmland birds in homogenized agricultural landscape: a cross-border comparison over the former Iron Curtain. Agric Ecosyst Environ. 2021;321:107628.
Tripathi BM, Edwards DP, Mendes LW, Kim M, Dong K, Kim H, et al. The impact of tropical forest logging and oil palm agriculture on the soil microbiome. Mol Ecol. 2016;25:2244–57.
Philippot L, Spor A, Hénault C, Bru D, Bizouard F, Jones CM, et al. Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 2013;7:1609–19.
Wang GH, Jin J, Liu JJ, Chen XL, Liu JD, Liu XB. Bacterial community structure in a mollisol under long-term natural restoration, cropping, and bare fallow history estimated by PCR-DGGE. Pedosphere. 2009;19:156–65.
Delgado-Baquerizo M, Eldridge DJ, Liu YR, Sokoya B, Wang JT, Hu HW, et al. Global homogenization of the structure and function in the soil microbiome of urban greenspaces. Sci Adv. 2021;7:eabg5809.
Hartmann FE, McDonald BA, Croll D. Genome-wide evidence for divergent selection between populations of a major agricultural pathogen. Mol Ecol. 2018;27:2725–41.
Hufnagel J, Reckling M, Ewert F. Diverse approaches to crop diversification in agricultural research. A review. Agron Sustain Dev. 2020;40:1–17.
Duan Y, Wang X, Wang L, Lian J, Wang W, Wu F, et al. Biogeographic patterns of soil microbe communities in the deserts of the Hexi Corridor, northern China. Catena. 2022;211:106026.
Zhu W, Zhu M, Liu X, Xia J, Yin H, Li X. Different responses of bacteria and microeukaryote to assembly processes and co-occurrence pattern in the coastal upwelling. Microb Ecol. 2022;86:174–86.
Gong X, Liu X, Li Y, Ma K, Song W, Zhou J, et al. Distinct ecological processes mediate domain-level differentiation in microbial spatial scaling. Appl Environ Microbiol. 2023;89:e02096–22.
Feng M, Tripathi BM, Shi Y, Adams JM, Zhu YG, Chu H. Interpreting distance‐decay pattern of soil bacteria via quantifying the assembly processes at multiple spatial scales. MicrobiologyOpen 2019;8:e00851.
Clark DR, Underwood GJ, McGenity TJ, Dumbrell AJ. What drives study‐dependent differences in distance-decay relationships of microbial communities? Glob Ecol Biogeogr. 2021;30:811–25.
Schadt CW, Chen H, Ma K, Lu C, Fu Q, Qiu Y, et al. Functional redundancy in soil microbial community based on metagenomics across the globe. Front Microbiol. 2022;13:87897.
Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci USA. 2011;108:14288–93.
Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci USA. 2012;109:21390–5.
Guan X, Wang J, Zhao H, Wang J, Luo X, Liu F, et al. Soil bacterial communities shaped by geochemical factors and land use in a less-explored area, Tibetan Plateau. BMC Genom. 2013;14:1–13.
Manoharan L, Kushwaha SK, Ahrén D, Hedlund K. Agricultural land use determines functional genetic diversity of soil microbial communities. Soil Biol Biochem. 2017;115:423–32.
Monteux S, Keuper F, Fontaine S, Gavazov K, Hallin S, Juhanson J, et al. Carbon and nitrogen cycling in Yedoma permafrost controlled by microbial functional limitations. Nat Geosci. 2020;13:794–8.
Aryal B, Gurung R, Camargo AF, Fongaro G, Treichel H, Mainali B, et al. Nitrous oxide emission in altered nitrogen cycle and implications for climate change. Environ Pollut. 2022;314:120272.
Mohapatra M, Yadav R, Rajput V, Dharne MS, Rastogi G. Metagenomic analysis reveals genetic insights on biogeochemical cycling, xenobiotic degradation, and stress resistance in mudflat microbiome. J Environ Manag. 2021;292:112738.
Merloti LF, Mendes LW, Pedrinho A, de Souza LF, Ferrari BM, Tsai SM. Forest to agriculture conversion in Amazon drives soil microbial communities and N-cycle. Soil Biol Biochem. 2019;137:107567.
Chen Y, Yang X, Fu W, Chen B, Hu H, Feng K, et al. Conversion of natural grassland to cropland alters microbial community assembly across northern China. Environ Microbiol. 2022;24:5630–42.
Jiao S, Xu Y, Zhang J, Hao X, Lu Y. Core microbiota in agricultural soils and their potential associations with nutrient cycling. Msystems 2019;4:e00313–18.
Jiao S, Chen W, Wei G. Core microbiota drive functional stability of soil microbiome in reforestation ecosystems. Glob Change Biol. 2022;28:1038–47.
Shi Q, Wang T, Lee C. MEMS based broadband piezoelectric ultrasonic energy harvester (PUEH) for enabling self-powered implantable biomedical devices. Sci Rep. 2016;6:1–10.
Liu X, Lee Burras C, Kravchenko YS, Duran A, Huffman T, Morras H, et al. Overview of mollisols in the world: distribution, land use and management. Can J Soil Sci. 2012;92:383–402.
Walsh MG, de Smalen AW, Mor SM. Climatic influence on anthrax suitability in warming northern latitudes. Sci Rep. 2018;8:9269.
Chendev YG, Sauer TJ, Hernandez Ramirez G, Burras CL. History of east European chernozem soil degradation; protection and restoration by tree windbreaks in the Russian steppe. Sustainability .2015;7:705–24.
Stakhurlova L, Svistova I, Shcheglov D. Biological activity as an indicator of chernozem fertility in different biocenoses. Eurasia Soil Sci. 2007;40:694–9.
Eremin D. Changes in the content and quality of humus in leached chernozems of the Trans-Ural forest-steppe zone under the impact of their agricultural use. Eurasia Soil Sci. 2016;49:538–45.
Lal R. Managing soils and ecosystems for mitigating anthropogenic carbon emissions and advancing global food security. BioScience 2010;60:708–21.
Zhang Y, Liang A, Wang Y, Chen X, Zhang S, Jia S, et al. Climate change impacts on soil fertility in Chinese Mollisols. In Sustainable Crop Productivity and Quality under Climate Change. Academic Press: Cambridge, MA, USA, 2022. pp 275–93.
Shi Y, Li Y, Xiang X, Sun R, Yang T, He D, et al. Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome. 2018;6:1–12.
Paul BG, Burstein D, Castelle CJ, Handa S, Arambula D, Czornyj E, et al. Retroelement-guided protein diversification abounds in vast lineages of Bacteria and Archaea. Nat Microbiol. 2017;2:1–7.
Zhang T, Qiao Q, Novikova PY, Wang Q, Yue J, Guan Y, et al. Genome of Crucihimalaya himalaica, a close relative of Arabidopsis, shows ecological adaptation to high altitude. Proc Natl Acad Sci USA. 2019;116:7137–46.
Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:1–9.
Anderson M PCO: a FORTRAN computer program for principal coordinate analysis. Department of Statistics, University of Auckland, New Zealand. 2003.
Anderson MJ Permutational multivariate analysis of variance. In Wiley StatsRef: Statistics Reference Online. Wiley: Hoboken, NJ, USA, 2017. pp 1–5.
Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22.
Chen QL, Xiang Q, Sun AQ, Hu HW. Aridity differentially alters the stability of soil bacterial and fungal networks. Environ Microbiol. 2022;24:5574–82.
Ma B, Stirling E, Liu Y, Zhao K, Zhou J, Singh BK, et al. Soil biogeochemical cycle couplings inferred from a function-taxon network. Research 2021;2021:7102769.
Ma B, Zhao K, Lv X, Su W, Dai Z, Gilbert JA, et al. Genetic correlation network prediction of forest soil microbial functional organization. ISME J. 2018;12:2492–505.
Delgado-Baquerizo M, Maestre FT, Reich PB, Jeffries TC, Gaitan JJ, Encinar D, et al. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat Commun. 2016;7:10541.
Flynn DF, Gogol-Prokurat M, Nogeire T, Molinari N, Richers BT, Lin BB, et al. Loss of functional diversity under land use intensification across multiple taxa. Ecol Lett. 2009;12:22–33.
Milder JC, Clark S. Conservation development practices, extent, and land-use effects in the United States. Conserv Biol. 2011;25:697–707.
Mayfield MM, Bonser S, Morgan J, Aubin I, McNamara S, Vesk P. What does species richness tell us about functional trait diversity? Predictions and evidence for responses of species and functional trait diversity to land-use change. Glob Ecol Biogeogr. 2010;19:423–31.
Farneda FZ, Grelle CE, Rocha R, Ferreira DF, López-Baucells A, Meyer CF. Predicting biodiversity loss in island and countryside ecosystems through the lens of taxonomic and functional biogeography. Ecography. 2020;43:97–106.
Trivedi C, Delgado-Baquerizo M, Hamonts K, Lai K, Reich PB, Singh BK. Losses in microbial functional diversity reduce the rate of key soil processes. Soil Biol Biochem. 2019;135:267–74.
Verbruggen E, Toby, Kiers E. Evolutionary ecology of mycorrhizal functional diversity in agricultural systems. Evol Appl. 2010;3:547–60.
Estel S, Kuemmerle T, Levers C, Baumann M, Hostert P. Mapping cropland-use intensity across Europe using MODIS NDVI time series. Environ Res Lett. 2016;11:024015.
Menta C Soil fauna diversity-function, soil degradation, biological indices, soil restoration. In: Lameed, GA (eds). Biodiversity Conservation and Utilization in a Diverse World. IntechOpen, London, UK, 2012. pp 59–94.
Prinzing A, Reiffers R, Braakhekke WG, Hennekens SM, Tackenberg O, Ozinga WA, et al. Less lineages–more trait variation: phylogenetically clustered plant communities are functionally more diverse. Ecol Lett. 2008;11:809–19.
Homburg JA, Sandor JA. Anthropogenic effects on soil quality of ancient agricultural systems of the American Southwest. Catena. 2011;85:144–54.
Crow WT, Berg AA, Cosh MH, Loew A, Mohanty BP, Panciera R, et al. Upscaling sparse ground-based soil moisture observations for the validation of coarse-resolution satellite soil moisture products. Rev Geophys. 2012;50:GT2002.
Frainer A, Primicerio R, Kortsch S, Aune M, Dolgov AV, et al. Climate-driven changes in functional biogeography of Arctic marine fish communities. Proc Natl Acad Sci USA. 2017;114:12202–7.
Rey A, Pegoraro E, Oyonarte C, Were A, Escribano P, Raimundo J. Impact of land degradation on soil respiration in a steppe (Stipa tenacissima L.) semi-arid ecosystem in the SE of Spain. Soil Biol Biochem. 2011;43:393–403.
He Z, Xu M, Deng Y, Kang S, Kellogg L, Wu L, et al. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett. 2010;13:564–75.
Su JQ, Ding LJ, Xue K, Yao HY, Quensen J, Bai SJ, et al. Long-term balanced fertilization increases the soil microbial functional diversity in a phosphorus-limited paddy soil. Mol Ecol. 2015;24:136–50.
Poeplau C, Don A, Vesterdal L, Leifeld J, Van Wesemael B, Schumacher J, et al. Temporal dynamics of soil organic carbon after land-use change in the temperate zone-carbon response functions as a model approach. Glob Change Biol. 2011;17:2415–27.
Bischoff N, Mikutta R, Shibistova O, Puzanov A, Reichert E, Silanteva M, et al. Land-use change under different climatic conditions: Consequences for organic matter and microbial communities in Siberian steppe soils. Agric, Ecosyst Environ. 2016;235:253–64.
Jacinthe P-A, Lal R. Labile carbon and methane uptake as affected by tillage intensity in a Mollisol. Soil Tillage Res. 2005;80:35–45.
Yuan H, Ge T, Chen X, Liu S, Zhu Z, Wu X, et al. Abundance and diversity of CO2-assimilating bacteria and algae within red agricultural soils are modulated by changing management practice. Micro Ecol. 2015;70:971–80.
Xiao KQ, Ge TD, Wu XH, Peacock CL, Zhu ZK, Peng J, et al. Metagenomic and 14C tracing evidence for autotrophic microbial CO2 fixation in paddy soils. Environ Microbiol. 2021;23:924–33.
Anderson-Teixeira KJ, Masters MD, Black CK, Zeri M, Hussain MZ, Bernacchi CJ, et al. Altered belowground carbon cycling following land-use change to perennial bioenergy crops. Ecosystems. 2013;16:508–20.
Yu L, Luo S, Xu X, Gou Y, Wang J. The soil carbon cycle determined by GeoChip 5.0 in sugarcane and soybean intercropping systems with reduced nitrogen input in South China. Appl Soil Ecol. 2020;155:103653.
Dobbelaere S, Vanderleyden J, Okon Y. Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit Rev Plant Sci. 2003;22:107–49.
Blaud A, van der Zaan B, Menon M, Lair GJ, Zhang D, Huber P, et al. The abundance of nitrogen cycle genes and potential greenhouse gas fluxes depends on land use type and little on soil aggregate size. Appl Soil Ecol. 2018;125:1–11.
Schmidt CS, Richardson DJ, Baggs EM. Constraining the conditions conducive to dissimilatory nitrate reduction to ammonium in temperate arable soils. Soil Biol Biochem. 2011;43:1607–11.
Gao W, Kou L, Yang H, Zhang J, Müller C, Li S. Are nitrate production and retention processes in subtropical acidic forest soils responsive to ammonium deposition? Soil Biol Biochem. 2016;100:102–9.
Stevens R, Laughlin R, Malone J. Soil pH affects the processes reducing nitrate to nitrous oxide and dinitrogen. Soil Biol Biochem. 1998;30:1119–26.
Zhang Y, Zheng X, Ren X, Zhang J, Misselbrook T, Cardenas L, et al. Land-use type affects nitrate production and consumption pathways in subtropical acidic soils. Geoderma .2019;337:22–31.
Zhu X, Burger M, Doane TA, Horwath WR. Ammonia oxidation pathways and nitrifier denitrification are significant sources of N2O and NO under low oxygen availability. Proc Natl Acad Sci USA. 2013;110:6328–33.
Liang M, Frank S, Lünsdorf H, Warren MJ, Prentice MB. Bacterial microcompartment‐directed polyphosphate kinase promotes stable polyphosphate accumulation in E. coli. Biotechnol J. 2017;12:1600415.
Richardson AE, Lynch JP, Ryan PR, Delhaize E, Smith FA, Smith SE, et al. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil. 2011;349:121–56.
Bergkemper F, Schöler A, Engel M, Lang F, Krüger J, Schloter M, et al. Phosphorus depletion in forest soils shapes bacterial communities towards phosphorus recycling systems. Environ Microbiol. 2016;18:1988–2000.
Yao Q, Li Z, Song Y, Wright SJ, Guo X, Tringe SG, et al. Community proteogenomics reveals the systemic impact of phosphorus availability on microbial functions in tropical soil. Nat Ecol Evol. 2018;2:499–509.
Zhao FJ, Tausz M, De Kok LJ Role of sulfur for plant production in agricultural and natural ecosystems. In: R Hell, C Dahl, D Knaff, T Leustek (eds). Sulfur metabolism in phototrophic organisms. Advances in photosynthesis and respiration, vol. 27. Dordrecht, the Netherlands, Springer, 2008. pp 417–35.
Edmeades D, Thorrold B, Roberts A. The diagnosis and correction of sulfur deficiency and the management of sulfur requirements in New Zealand pastures: a review. Aust J Exp Agric. 2005;45:1205–23.
Wang F, Liang Y, Jiang Y, Yang Y, Xue K, Xiong J, et al. Planting increases the abundance and structure complexity of soil core functional genes relevant to carbon and nitrogen cycling. Sci Rep. 2015;5:14345.
Xie H, Huang Y, Chen Q, Zhang Y, Wu Q. Prospects for agricultural sustainable intensification: a review of research. Land. 2019;8:157.
Trivedi P, Mattupalli C, Eversole K, Leach JE. Enabling sustainable agriculture through understanding and enhancement of microbiomes. N. Phytol. 2021;230:2129–47.
Liu M, Zhang G, Yin F, Wang S, Li L. Relationship between biodiversity and ecosystem multifunctionality along the elevation gradient in alpine meadows on the eastern Qinghai-Tibetan plateau. Ecol Indic. 2022;141:109097.
Morris BE, Henneberger R, Huber H, Moissl-Eichinger C. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev. 2013;37:384–406.
Funding
This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA28020201, XDA28070302), Key Research Program of Frontier Sciences, Chinese Academy of Sciences (ZDBS-LY-DQC017), National Key Research and Development Program of China (2022YFD1500200), National Science and Technology Basic Resources Investigation Special Project (2021FY100400), National Natural Science Foundation of China (42177105), CAS International partnership project (131323KYSB20210004), Natural Science Foundation of Heilongjiang Province (ZD2022D001), and Youth Innovation Promotion Association of Chinese Academy of Sciences (2023237).
Author information
Authors and Affiliations
Contributions
JJL, JMA and GHW conceptualized and managed the study. JJL, LJL, ZHY, YSL and ZXL generated the data. HDG, ZXL, YPG and XJH analyzed and interpreted the data. JJL, JMA, YPG, and GHW drafted the manuscript. ZXL, JMA, JJ, YYS and XBL reviewed and edited the corrected manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, J., Guo, Y., Gu, H. et al. Conversion of steppe to cropland increases spatial heterogeneity of soil functional genes. ISME J 17, 1872–1883 (2023). https://doi.org/10.1038/s41396-023-01496-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-023-01496-9
This article is cited by
-
Metagenomic data highlight shifted nitrogen regime induced by wetland reclamation
Biology and Fertility of Soils (2024)