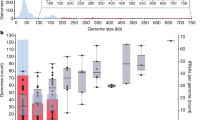
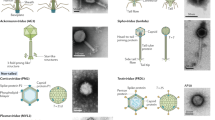
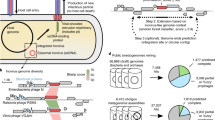
Abstract
Phages are prevalent in diverse environments and play major ecological roles attributed to their tremendous diversity and abundance. Among these viruses, transposable phages (TBPs) are exceptional in terms of their unique lifestyle, especially their replicative transposition. Although several TBPs have been isolated and the life cycle of the representative phage Mu has been extensively studied, the diversity distribution and ecological functions of TBPs on the global scale remain unknown. Here, by mining TBPs from enormous microbial genomes and viromes, we established a TBP genome dataset (TBPGD), that expands the number of accessible TBP genomes 384-fold. TBPs are prevalent in diverse biomes and show great genetic diversity. Based on taxonomic evaluations, we propose the categorization of TBPs into four viral groups, including 11 candidate subfamilies. TBPs infect multiple bacterial phyla, and seem to infect a wider range of hosts than non-TBPs. Diverse auxiliary metabolic genes (AMGs) are identified in the TBP genomes, and genes related to glycoside hydrolases and pyrimidine deoxyribonucleotide biosynthesis are highly enriched. Finally, the influences of TBPs on their hosts are experimentally examined by using the marine bacterium Shewanella psychrophila WP2 and its infecting transposable phage SP2. Collectively, our findings greatly expand the genetic diversity of TBPs, and comprehensively reveal their potential influences in various ecosystems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All the identified TBP genomic sequences (n = 18,449) have been deposited in CyVerse (available at https://data.cyverse.org/dav-anon/iplant/home/zhangmujie/TBPGD/TBPGD.zip) and in the National Omics Data Encyclopedia (NODE) under project ID OEP003495. The transcriptomic data from the current study have been deposited in NODE under project ID OEP002984.
References
Dion MB, Oechslin F, Moineau S. Phage diversity, genomics and phylogeny. Nat Rev Microbiol. 2020;18:125–38.
Zimmerman AE, Howard-Varona C, Needham DM, John SG, Worden AZ, Sullivan MB, et al. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat Rev Microbiol. 2020;18:21–34.
Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol. 2015;13:641–50.
Touchon M, Bernheim A, Rocha EP. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016;10:2744–54.
Wahl A, Battesti A, Ansaldi M. Prophages in Salmonella enterica: a driving force in reshaping the genome and physiology of their bacterial host? Mol Microbiol. 2019;111:303–16.
Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 2017;11:1511–20.
Argov T, Azulay G, Pasechnek A, Stadnyuk O, Ran-Sapir S, Borovok I, et al. Temperate bacteriophages as regulators of host behavior. Curr Opin Microbiol. 2017;38:81–7.
Toussaint A, Rice PA. Transposable phages, DNA reorganization and transfer. Curr Opin Microbiol. 2017;38:88–94.
Harshey RM. Transposable Phage Mu. Microbiology spectrum. 2014;2. https://doi.org/10.1128/microbiolspec.MDNA3-0007-2014.
Taylor AL. Bacteriophage-induced mutation in Escherichia coli. PNAS 1963;50:1043–51.
Harshey RM. The Mu story: how a maverick phage moved the field forward. Mobile. DNA 2012;3:21.
Mizuno N, Dramićanin M, Mizuuchi M, Adam J, Wang Y, Han Y-W, et al. MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition. PNAS 2013;110:E2441–50.
George M, Bukhari AI. Heterogeneous host DNA attached to the left end of mature bacteriophage Mu DNA. Nature 1981;292:175–6.
Groenen MAM, Putte PVD. Mapping of a site for packaging of bacteriophage Mu DNA. Virology 1985;144:520–2.
Howe MM. Transduction by Bacteriophage MU-l. Virology. 1973;55:103–17.
Gill GS, Hull RC. Mutator bacteriophage D108 and its DNA: an electron microscopic characterization. J Virol. 1981;37:420–30.
Braid MD, Silhavy JL, Kitts CL, Cano RJ, Howe MM. Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J Bacteriol. 2004;186:6560–74.
Summer EJ, Gonzalez CF, Carlisle T, Mebane LM, Cass AM, Savva CG, et al. Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. J Mol Biol. 2004;340:49–65.
Fogg PCM, Hynes AP, Digby E, Lang AS, Beatty JT. Characterization of a newly discovered Mu-like bacteriophage, RcapMu, in Rhodobacter capsulatus strain SB1003. Virology 2011;421:211–21.
Zehr ES, Tabatabai LB, Bayles DO. Genomic and proteomic characterization of SuMu, a Mu-like bacteriophage infecting Haemophilus parasuis. BMC Genom. 2012;13:331.
Jakhetia R, Verma NK. Identification and Molecular Characterisation of a Novel Mu-Like Bacteriophage, SfMu, of Shigella flexneri. PLoS ONE. 2015;10:e0124053.
Wu H, Zhang Y, Jiang Y, Wu H, Sun W, Huang Y-P. Characterization and Genomic Analysis of ΦSHP3, a New Transposable Bacteriophage Infecting Stenotrophomonas maltophilia. J Virol. 2021;95:e00019–21.
Masignani V, Giuliani MM, Tettelin H, Comanducci M, Rappuoli R, Scarlato V. Mu-like Prophage in serogroup B Neisseria meningitidis coding for surface-exposed antigens. Infect Immun. 2001;69:2580–8.
Morgan GJ, Hatfull GF, Casjens S, Hendrix RW. Bacteriophage Mu Genome Sequence Analysis and comparision with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J Mol Microbiol. 2002;317:337–59.
Guo Q, Chen B, Tu Y, Du S, Chen X. Prophage LambdaSo uses replication interference to suppress reproduction of coexisting temperate phage MuSo2 in Shewanella oneidensis MR-1. Environ Microbiol. 2019;21:2079–94.
Tang K, Lin D, Zheng Q, Liu K, Yang Y, Han Y, et al. Genomic, proteomic and bioinformatic analysis of two temperate phages in Roseobacter clade bacteria isolated from the deep-sea water. BMC Genom. 2017;18:485.
Szafrański SP, Kilian M, Yang I. Wieden GBd, Winkel A, Hegermann J, et al. Diversity patterns of bacteriophages infecting Aggregatibacter and Haemophilus species across clades and niches. ISME J. 2019;13:2500–22.
Cui Z, Xu Z, Wei Y, Zhang Q, Qin K, Ji X. Characterization and Genome Analysis of a Novel Mu-like Phage VW-6B Isolated from the Napahai Plateau Wetland of China. Curr Microbiol. 2021;78:150–8.
Lin D, Tang K, Han Y, Li C, Chen X. Genome sequence of an inducible phage in Rhodovulum sp. P5 isolated from the shallow-sea hydrothermal system. Mar Genom. 2016;30:93–5.
Mara P, Vik D, Pachiadaki MG, Suter EA, Poulos B, Taylor GT, et al. Viral elements and their potential influence on microbial processes along the permanently stratified Cariaco Basin redoxcline. ISME J. 2020;14:3079–92.
Hulo C, Masson P, Mercier PL, Toussaint A. A structured annotation frame for the transposable phages: a new proposed family “Saltoviridae” within the Caudovirales. Virology 2015;477:155–63.
Toussaint A, Gijsegem FV. Extension of the transposable bacterial virus family: two genomic organisations among phages and prophages with a Tn552-related transposase. Res Microbiol. 2018;169:495–9.
Ndela EO, Enault F, Toussaint A. Transposable Prophages in Leptospira: An Ancient, Now Diverse, Group Predominant in Causative Agents of Weil’s Disease. Int J Mol Sci. 2021;22:13434.
Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010;38:4207–17.
Paez-Espino D, Roux S, Chen I-MA, Palaniappan K, Ratner A, Chu K, et al. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res. 2019;47:D678–D86.
Gregory AC, Zayed AA, Conceição-Neto N, Temperton B, Bolduc B, Alberti A, et al. Marine DNA Viral Macro- and Microdiversity from Pole to Pole. Cell 2019;177:1109–23.
Turner D, Kropinski AM, Adriaenssens EM. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021;13:506.
Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil P-A, Hugenholtz P. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022;50:D785–D94.
Ritacco CJ, Kamtekar S, Wang J, Steitz TA. Crystal structure of an intermediate of rotating dimers within the synaptic tetramer of the G-segment invertase. Nucleic Acids Res. 2013;41:2673–82.
Howe MM, Pato ML. Phage Mu. Reference Module in Life Sciences. 2017:1–6. https://doi.org/10.1016/B978-0-12-809633-8.06883-7.
Shaffer M, Borton MA, McGivern BB, Zayed AA, Rosa SLL, Solden LM, et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 2020;48:8883–900.
Xiao X, Wang P, Zeng X, Bartlett DH, Wang F. Shewanella psychrophila sp. nov. and Shewanella piezotolerans sp. nov., isolated from west Pacific deep-sea sediment. Int J Syst Evolut Microbiol. 2007;57:60–5.
Xu G, Jian H, Xiao X, Wang F. Complete genome sequence of Shewanella psychrophila WP2, a deep-sea bacterium isolated from west Pacific sediment. Mar Genom. 2017;35:19–21.
Namgoong S-Y, M.Harshey R. The same two monomers within a MuA tetramer provide the DDE domains for the strand cleavage and strand transfer steps of transposition. EMBO J. 1988;17:3775–85.
Mizuuchi M, Mizuuchi K. Conformational isomerization in phage Mu transpososome assembly effects of the transpositional enhancer and of MuB. EMBO J. 2001;20:6927–35.
Han Y-W, Mizuuchi K. Phage Mu transposition immunity: protein pattern formation along DNA by a diffusion-ratchet mechanism. Mol Cell. 2010;39:48–58.
Choi W, Jang S, Harshey RM. Mu transpososome and RecBCD nuclease collaborate in the repair of simple Mu insertions. PNAS 2014;111:14112–7.
Wang PW, Chu L, Guttman DS. Complete sequence and evolutionary genomic analysis of the Pseudomonas aeruginosa transposable bacteriophage D3112. J Bacteriol. 2004;186:400–10.
Goudie AD, Lynch KH, Seed KD, Stothard P, Shrivastava S, Wishart DS, et al. Genomic sequence and activity of KS10, a transposable phage of the Burkholderia cepacia complex. BMC Genom. 2008;9:615.
Chung I-Y, Cho Y-H. Complete genome sequences of two Pseudomonas aeruginosa temperate phages, MP29 and MP42, which lack the phage-host CRISPR interaction. J Virol. 2012;86:8336.
Yang J, Kong Y, Li X, Yang S. A novel transposable Mu-like prophage in Bacillus alcalophilus CGMCC 1.3604 (ATCC 27647). Virol Sin. 2015;30:63–5.
Thi BVT, Khanh NHP, Namikawa R, Miki K, Kondo A, Thi PTD, et al. Genomic characterization of Ralstonia solanacearum phage ΦRS138 of the family Siphoviridae. Arch Virol. 2016;161:483–6.
Cornuault JK, Petit M-A, Mariadassou M, Benevides L, Moncaut E, Langella P, et al. Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome 2018;6:65.
Toussaint A. Transposable Mu-like phages in Firmicutes: new instances of divergence generating retroelements. Res Microbiol. 2013;164:281–7.
Cazares A, Mendoza-Hernández G, Guarneros G. Core and accessory genome architecture in a group of Pseudomonas aeruginosa Mu-like phages. BMC Genom. 2014;15:1146.
Touchon M, Sousa JAMD, Rocha EP. Embracing the enemy: the diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr Opin Microbiol. 2017;38:66–73.
Edlin G, Lin L, Bitner R. Reproductive fitness of P1, P2, and Mu lysogens of Escherichia coli. J Virol. 1977;21:560–4.
Engelhardt T, Sahlberg M, Cypionka H, Engelen B. Biogeography of Rhizobium radiobacter and distribution of associated temperate phages in deep subseafloor sediments. ISME J. 2013;7:199–209.
Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, et al. Towards environmental systems biology of Shewanella. Nat Rev Microbiol. 2008;6:592–603.
Lemaire ON, Méjean V, Iobbi-Nivol C. The Shewanella genus: ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol Rev. 2020;44:155–70.
Hickman AB, Dyda F. DNA Transposition at Work. Chem Rev. 2016;116:12758–84.
Roux S, Krupovic M, Daly RA, Borges AL, Nayfach S, Schulz F, et al. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes. Nat Microbiol. 2019;4:1895–906.
Sayers EW, Cavanaugh M, Clark K, Pruitt KD, Schoch CL, Sherry ST, et al. GenBank. Nucleic Acids Res. 2021;49:D92–D6.
Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018;46:D708–D17.
Steinegger M, Meier M, Mirdita M, Vöhringer H, Haunsberger SJ, Söding J. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinform. 2019;20:473.
Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D9.
Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE. 2014;9:e108277.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–9.
Holm L. Using Dali for Protein Structure Comparison. In: Gάspάri Z, editor. Structural Bioinformatics: Methods and Protocols. Methods in Molecular Biology. Springer Science+Business Media; 2020;2112:29–42.
Li W, O’Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, et al. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021;49:D1020–D8.
O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–45.
Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40:D26–32.
Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, et al. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014;42:D26–31.
Bolduc B, Zablocki O, Guo J, Zayed AA, Vik D, Dehal P, et al. iVirus 2.0: Cyberinfrastructure-supported tools and data to power DNA virus ecology. ISME Commun. 2021;1:77.
Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119.
Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–W4.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421.
Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60.
Terzian P, Ndela EO, Galiez C, Lossouarn J, Bucio REP, Mom R, et al. PHROG: families of prokaryotic virus proteins clustered using remote homology. NAR Genom Bioinforma. 2021;3:lqab067.
Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 2012;28:3150–2.
Roux S, Adriaenssens EM, Dutilh BE, Koonin EV, Kropinski AM, Krupovic M, et al. Minimum Information about an Uncultivated Virus Genome (MIUViG). Nat Biotechnol. 2019;37:29–37.
Jang HB, Bolduc B, Zablocki O, Kuhn JH, Roux S, Adriaenssens EM, et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat Biotechnol. 2019;37:632–9.
Shannon P, Andrew M, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–504.
Jiang J-Z, Yuan W-G, Shang J, Shi Y-H, Yang L-L, Liu M, et al. Virus classification for viral genomic fragments using PhaGCN2. Brief Bioinforma. 2023;24:1–9.
Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020;36:1925–7.
Matsen FA, Kodner RB, Armbrust EV. pplacer: linear time maximum-likelihood and bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinform. 2010;11:538.
Consortium TU. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D9.
Yan F, Yu X, Duan Z, Lu J, Jia B, Qiao Y, et al. Discovery and characterization of the evolution, variation and functions of diversity-generating retroelements using thousands of genomes and metagenomes. BMC Genom. 2019;20:595.
Jian H, Yi Y, Wang J, Hao Y, Zhang M, Wang S, et al. Diversity and distribution of viruses inhabiting the deepest ocean on Earth. ISME J. 2021;15:3094–110.
Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinform. 2007;8:209.
Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, et al. Uncovering Earth’s virome. Nature 2016;536:425–30.
Guo J, Bolduc B, Zayed AA, Varsani A, Dominguez-Huerta G, Delmont TO, et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome 2021;9:37.
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D51.
Chan PP, Lin BY, Mak AJ, Lowe TM. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021;49:9077–96.
Ahlmann-Eltze C, Patil I. Ggsignif: R package for displaying significance brackets for ‘ggplot2’. PsyArXiv. 2021. https://doi.org/10.31234/osf.io/7awm6.
Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–72.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40.
Benjamini Y, Hochberg Y. Controlling the False Discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B (Methodol). 1995;1:289–300.
Meng C, Li S, Fan Q, Chen R, Hu Y, Xiao X, et al. The thermo-regulated genetic switch of deep-sea filamentous phage SW1 and its distribution in the Pacific Ocean. FEMS Microbiol Lett. 2020;367:fnaa094.
Hurwitz BL, Sullivan MB. The Pacific Ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS ONE. 2013;8:e57355.
Jian H, Xiao X, Wang F. Role of filamentous phage SW1 in regulating the lateral flagella of Shewanella piezotolerans strain WP3 at low temperatures. Appl Environ Microbiol. 2013;79:7101–9.
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134.
Jian H, Xu G, Yi Y, Hao Y, Wang Y, Xiong L, et al. The origin and impeded dissemination of the DNA phosphorothioation system in prokaryotes. Nat Commun. 2021;12:6382.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018;34:i884–i90.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323.
Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010;26:136–8.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Baren MJV, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5.
Acknowledgements
This work was financially supported by the Hainan Provincial Joint Project of Sanya Yazhou Bay Science and Technology City (grant no. 2021JJLH0057), the National Natural Science Foundation of China (grant nos. 42176095, 91851113, 41921006), the National Key R&D Program of China (grant no. 2021YFF0501300), and the Oceanic Interdisciplinary Program of Shanghai Jiao Tong University (project no. SL2021PT201). We would like to thank Prof. Ariane Toussaint for helpful suggestions on TBP identification. We are grateful to the editor and two anonymous reviewers for their comments that were instrumental in improving the paper.
Author information
Authors and Affiliations
Contributions
HJ conceived and designed the research; MZ, QS, and ST collected and curated genomic and metadata; MZ performed the bioinformatic and statistical analysis; MZ and SL conducted the microbiological experiments; YH and XT helped in RNA isolation; MZ and HJ analyzed and interpreted the data; HJ and MZ wrote the paper; XX and YY provided useful comments to improve the paper; HJ supervised the project. All the authors reviewed the results and approved the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, M., Hao, Y., Yi, Y. et al. Unexplored diversity and ecological functions of transposable phages. ISME J 17, 1015–1028 (2023). https://doi.org/10.1038/s41396-023-01414-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-023-01414-z