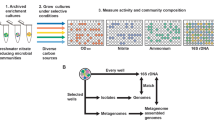
Abstract
Small regulatory RNAs and antisense RNAs play important roles in the regulation of gene expression in bacteria but are underexplored, especially in natural populations. While environmentally relevant microbes often are not amenable to genetic manipulation or cannot be cultivated in the laboratory, extensive metagenomic and metatranscriptomic datasets for these organisms might be available. Hence, dedicated workflows for specific analyses are needed to fully benefit from this information. Here, we identified abundant sRNAs from oceanic environmental populations of the ecologically important primary producer Prochlorococcus starting from a metatranscriptomic differential RNA-Seq (mdRNA-Seq) dataset. We tracked their homologs in laboratory isolates, and we provide a framework for their further detailed characterization. Several of the experimentally validated sRNAs responded to ecologically relevant changes in cultivation conditions. The expression of the here newly discovered sRNA Yfr28 was highly stimulated in low-nitrogen conditions. Its predicted top targets include mRNAs encoding cell division proteins, a sigma factor, and several enzymes and transporters, suggesting a pivotal role of Yfr28 in the coordination of primary metabolism and cell division. A cis-encoded antisense RNA was identified as a possible positive regulator of atpF encoding subunit b’ of the ATP synthase complex. The presented workflow will also be useful for other environmentally relevant microorganisms for which experimental validation abilities are frequently limiting although there is wealth of sequence information available.
Similar content being viewed by others
Introduction
Non-coding (nc)RNAs such as trans-acting small (s)RNAs and antisense RNAs (asRNAs) that overlap with other transcripts in cis are important components in the global control of gene expression in bacteria. Their functional roles have been studied in detail in genetically tractable bacteria such as Escherichia coli, Bacillus subtilis, Staphylococcus aureus, Pseudomonas aeruginosa, and Vibrio cholera (for reviews see [1,2,3,4,5]). However, there are many other important groups of bacteria for which no genetic tools exist or which even cannot be cultivated in the laboratory, enormously hampering the exploration of their respective sets of regulatory RNAs. On the other hand, especially for environmentally relevant microorganisms, frequently vast quantities of sequence data are available from extensive metagenomic and metatranscriptomic surveys, and such information can be expected to become even more readily available in the future.
The marine cyanobacterium Prochlorococcus is the most abundant phototroph throughout the euphotic zone of the vast oligotrophic areas of the oceans between 40°N and 40°S [6]. On the basis of extrapolations, it has been calculated that there are a total of 1027 Prochlorococcus cells on Earth, which fix an estimated four gigatons of carbon per year, comparable to the net primary production of all croplands in the world [7]. Phylogenetically distinct ecotypes of Prochlorococcus inhabit the oceans [8], which can be divided into two groups according to their adaptation to low light (LL) or high light (HL) conditions [9]. At the northern tip of the Gulf of Aqaba (Station A, 29°28′N 34°55′E) - the sampling site of this study – Prochlorococcus cells can reach a density of up to 2 × 105 per ml during the summer at the height of stratification [10] and are dominated by HL clade II (eMIT9312) and LL clade noncultured 1 (NC1), which is most closely related to LL I (eNATL2a) [11].
Prochlorococcus genome sizes vary between 1.62 and 2.68 Mbp [12], which is very small for a free-living photoautotroph. The highly streamlined genomes of Prochlorococcus have been interpreted as an adaptation to ultralow nutrient conditions [13]. The reduced genome size correlates with a small number of protein regulators [8]. Therefore, the control of gene expression by sRNA regulators might have been particularly important during the evolution of this phylum. Indeed, a relatively large number of sRNAs have been found in Prochlorococcus by computational prediction, microarray analysis and high-throughput sequencing [14,15,16]. However, despite the wealth of available genome information, the knowledge of the transcriptional architecture and the numbers and types of potential regulatory RNA molecules is still largely fragmentary and limited to the HL-adapted MED4 strain [15] and the LL-adapted MIT9313 strain [16].
We therefore collected seawater samples from three different depths in the Gulf of Aqaba, Red Sea, and extracted RNA for RNA-seq. The sampling location was chosen because it is (i) well characterized, (ii) has been monitored for decades (including physicochemical properties and the planktonic community composition) and (iii) is well known for the high abundance of Prochlorococcus during summer, allowing the straightforward collection of sufficient quantities of cells.
The identification of key RNA regulators within microbial communities is still challenging, and a convenient pipeline for in silico analysis and experimental verification has not yet been deployed. Here, we present a computational workflow for the identification of putative sRNAs based on the analysis of environmental transcriptome datasets and their experimental validation. These analyses led to the discovery of several new Prochlorococcus-specific sRNAs, which are likely of ecological importance.
Materials and methods
Preprocessing and global read assignment
For the identification of new sRNAs, we used three datasets from samples that were collected at station A in the Red Sea at depths of 60, 100, and 130 m [17]. The sequencing and preprocessing of the reads included quality control, quality trimming, and removal of ribosomal RNA reads, as described previously [17]. The preprocessed datasets were aligned to the NCBI nt database using discontiguous MegaBLAST [18].
We utilized MEGAN’s built-in “LCA” (Lowest Common Ancestor) function to achieve a higher taxonomic assignment resolution by reassigning multimapped reads to a higher taxonomic level [19]. Analyses were performed with the default settings. For normalization and selection of taxa of interest, we extracted global taxonomic tree information (number of assigned reads per taxon) from MEGAN [20]. Based on the three datasets, we computed a comparative weighted Venn tree for the Cyanobacteria phylum focusing on Prochlorococcus with CoVennTree v1.6.0 [20].
Computational identification of sRNA candidates
From the MEGAN output, we extracted Prochlorococcus-related reads in multi-FASTA format. For the computational analysis of sRNA candidates, we merged the three multi-FASTA datasets (60, 100, and 130 m) into a single file, which was further condensed into contigs using the de novo assembler Trinity v2.1.1 [21]. Next, the contigs were mapped against the well-annotated genomes of two Prochlorococcus model strains, MED4 (accession number: BX548174) and NATL2A (accession number: CP000095), using segemehl [22, 23]. Only contigs that did not overlap at all with a protein-coding gene were kept, and these contigs were used as the input to search for homologs throughout the entire cyanobacterial phylum using the GLASSgo v1.220 algorithm [24]. The putative sRNA homologs predicted by GLASSgo were aligned with Clustal Omega v1.2.3 and scored with RNAz v2.1, which computes a z-score based on secondary structure conservation and thermodynamic stability [25, 26], using the default settings for both the Clustal Omega and RNAz. Trinity results (number of reads per contig), and the GLASSgo outputs in conjunction with the RNAz predictions were compiled into master tables (Tables S2, S3 for NATL2A and MED4, respectively).
Culturing, RNA preparation and northern blot analysis
Prochlorococcus NATL2A and MED4 cultures were grown at 22 °C in AMP1 medium [27] under 10–30 µmol quanta m−2 s−1 of continuous white cool light to cell densities of 1–3 × 108 cells per ml. Stress experiments were performed as described previously [16]. In brief, the cells were subjected to several stress conditions for 30 min: light stress (light shifts from 10 µE to 100 µE (NATL2A) or 30 µE to 300 µE (MED4) or darkness) and temperature stress (shifts from 22 °C to 12 °C or 32 °C, respectively). For nitrogen starvation, the cells were washed twice in nitrogen-free medium and grown in minus N medium for 2 days. Iron depletion was induced via the addition of 0.6 µM DFB 2 days before sampling. The cells were harvested via filtration onto Supor-450 membranes, snap frozen in liquid nitrogen in tubes containing 2 ml of PGTX buffer [28] and subsequently stored at – 80 °C. Total RNA was extracted following the hot phenol method [29]. Northern hybridizations were performed as described previously [30] using 5 µg (small polyacrylamide gels and denaturing agarose gels), 10 µg (large polyacrylamide gels) or 50 µg (identification of new sRNAs on large polyacrylamide gels) of total RNA per sample. The primers used for the generation of specific probes are listed in Table S1.
Primer extension
Transcript templates for in vitro RNA synthesis were generated from purified PCR products or annealed complementary oligonucleotides using primers #17–20 (Table S1). The desired RNAs were transcribed using a MEGAshortscript Kit (ThermoFisher Scientific), and residual DNA was removed by TURBO DNase I treatment, with both steps performed according to the manufacturer’s instructions. RNA was purified using RNA Clean & Concentrator columns (Zymo Research) following the manufacturer’s instructions. If required, in vitro-transcribed RNA was separated on 7 M urea-10% polyacrylamide gels or on 2% nondenaturing agarose gels, and full-length fragments were excised and purified using either a ZR small-RNA PAGE Recovery Kit (Zymo Research) or a NucleoSpin Gel and PCR-Clean-up kit (Macherey-Nagel) according to the manufacturer’s instructions. The ppc_RT primer (#21, Table S1) was labeled as described previously [30]. Annealing mixtures containing 0.2 pmol of in vitro-synthesized ppc target RNA and 2 pmol of the 5′ end-labeled primer #21 (Table S1) without or with 40/80/160 pmol of in vitro-synthesized sRNA Yfr28 or 160 pmol of in vitro-synthesized sRNA Yfr2 were heated for 10 min at 70 °C and then chilled on ice for at least 5 min. cDNA synthesis was performed for 2 h at 30 °C using SuperScript III Reverse Transcriptase (ThermoFisher Scientific) according to the manufacturer’s instructions. The reaction was inactivated by incubation for 15 min at 70 °C, followed by RNase H treatment for 20 min at 37 °C and a final heat inactivation step of 5 min at 95 °C in RNA loading buffer. DNA sequencing ladder reactions were performed with the same 5′ end-labeled primer used for cDNA synthesis and the same template DNA employed for the in vitro synthesis of the target RNA using a USB Thermo Sequenase Cycle Sequencing Kit (Affymetrix). The primer extension products and sequencing reactions were separated on 8.3 M urea-6% polyacrylamide sequencing gels, and the vacuum-dried gels were exposed to imaging plates. Signals were visualized using a Typhoon FLA 9500 instrument (GE Healthcare) with Quantity One software (Bio-Rad).
Results and discussion
Workflow for the identification and characterization of sRNAs in environmentally relevant bacteria
Environmental samples were collected at Station A in the northern Gulf of Aqaba, Red Sea [17]. During the preparation of these samples, the RNA was treated with terminator exonuclease prior to library preparation [17] according to the differential RNA-Seq protocol [31]. This modification has several advantages, as it reduces the number of ribosomal RNAs and other processed transcripts, allows the precise mapping of transcriptional start sites, yields specific information about sRNAs and has been demonstrated to work well on environmental samples [32]. Here, all the sequenced reads from the samples obtained from the three different depths (60, 100, and 130 m) were processed as summarized in the workflow in Fig. 1. After taxonomic read assignment, we focused on the reads that could be assigned to genomic sequences belonging to the genus Prochlorococcus. These constituted 14.6, 8.1, and 0.7% of all reads from 60, 100, and 130 m. The known classification of Prochlorococcus according to ecotype was clearly visible in the mapping of the metatranscriptomic reads to distinct genomic sequences. Whereas the majority of Prochlorococcus-specific reads from 60 m mapped to Prochlorococcus of the HL ecotype, such as the AS9601, MIT9301, MIT9312, MIT9202, MIT9215, MED4, and MIT9515 strains, most of the reads from the two greater depths mapped to representatives of the LL ecotypes, such as the NATL1A, NATL2A, MIT9211, and SS120 strains (Fig. 2). Similar distribution profiles were detected by Shibl et al. [11] based on 16S–23 S rRNA internal transcribed spacer clone libraries that were generated from samples collected in September and October 2011 throughout the water column at the northern and southern ends of the Red Sea.
Samples were collected from 60 m (blue circles), 100 m (red circles), and 130 m (yellow circles) water depths. After the initial bioinformatic analysis (see workflow in Fig. 1), the weighted Venn tree was computed using CoVennTree [20]. The numbers in parentheses refer to weighted Venn decomposition values. To enable comparisons, each library was normalized to 10,000,000 reads. Of the entire computed CoVennTree graph only the cyanobacterial phylum is shown.
The next steps of the analysis included reassignment of the Prochlorococcus-related reads against the well-annotated genome sequences of two Prochlorococcus model strains, NATL2A (Files S1 and S2) [33] and MED4 [34]. We choose the LL strain NATL2A because this was one of the strains to which most of the reads could be assigned to within the LL clade, and the HL strain MED4 because this is the strain for which most information on Prochlorococcus sRNAs is available. The reads that mapped to intergenic regions (IGRs) and to ncRNAs (including housekeeping genes such as tRNAs, rnpB, ffs, and yfrs) were collected, unified and used as the input to search for homologs throughout the cyanobacterial phylum using the GLASSgo algorithm [24], which yielded 389 and 982 IGRs and annotated ncRNAs for NATL2A and MED4 (Tables S2 and S3). GLASSgo homologs were subjected to RNAz, which scores multiple sequence alignments of sRNA candidates based on secondary structure conservation and thermodynamic stability [25]. Based on the z-score (≤ −1), a total of 104 and 152 IGRs in NATL2A and MED4 were considered potential sRNA candidates (Tables S2, S3). Among these candidates, 34 in NATL2A and 42 in MED4 were previously annotated as tRNAs, housekeeping RNAs such as tmRNA and ffs, yfrs or ribosomal RNAs (Tables S2, S3). Many of the RNA classes were also found when searching in the RFAM database [35, 36] (Tables S2, S3). In total, we detected 24 of the 30 previously identified Prochlorococcus sRNAs [14,15,16] in the three datasets (Table S5) suggesting that our workflow is very suitable for the discovery of ncRNAs. The phylogenetic distribution of the MED4 and NATL2A sRNA candidates in other Prochlorococcus clades, cyanobacteria and other bacteria is given in Table S4.
Following manual inspection, selected sRNA candidates were experimentally validated and characterized in more detail, and sets of homologous sRNAs were subjected to target prediction in parallel (Tables S6, S7) using the CopraRNA package [37, 38]. The results will be presented in the following section.
Validation and characterization of predicted sRNAs and asRNAs in laboratory isolates
After the manual inspection of potential sRNAs, the most promising candidates were subjected to northern hybridization, and five of the tested candidates could be validated and showed typical sizes and structures (Fig. 3). In a recent study, we observed the Prochlorococcus HL and LL clade-specific occurrence of sRNAs [16]. Except for Yfr29, which was only present in MED4, all other newly detected sRNAs were Prochlorococcus- or even clade-specific, confirming our previous findings [16]. Because of their mode of action, sRNAs are often coregulated with the environmental conditions in which they play a role. With the exception of the MED4-specific sRNA Yfr29, all other sRNAs responded to tested environmental fluctuations such as variations in light intensity (Yfr107) and changes in temperature (Yfr28 and Yfr108), with the strongest responses being observed under nitrogen deprivation (Yfr28) and in the stationary phase (Yfr106) (Fig. 4). Interestingly, these were the same highly responsive conditions that we observed in our previous study [16]. Subsequently, we focused on the 72 nt-long sRNA Yfr28, which occurred in both subclades HLI (MED4 and MIT9515) and HLII (MIT9215, MIT9301, MIT9312, AS9601, and MIT0604). The yfr28 gene is framed on the opposite strand by the ftsQ and ftsZ genes, encoding cell division proteins (Fig. 5d). The synteny is highly conserved (Fig. S1). Next, we predicted targets for Yfr28 using CopraRNA [37, 38] including all 7 Prochlorococcus strains with a Yfr28 homolog (Table S6). First, to gain a better understanding of the interaction of Yfr28 with its targets, we investigated the temporal expression kinetics of Yfr28 during nitrogen-limiting conditions (Fig. 5a). The expression of Yfr28 was induced sevenfold after 3 h of nitrogen depletion and continuously increased to almost 200-fold of the initial expression value within 72 h (Fig. 5a). This is the most strongly induced Prochlorococcus-related sRNA identified in response to nitrogen starvation to date. In a previous study, we observed a tenfold increase in response to nitrogen limitation for the highly conserved and highly abundant sRNA Yfr2 [39]. The expression profiles are quite similar for Yfr28 and Yfr2; however, the latter sRNA reached its maximum level after 48 h, whereas Yfr28 had not yet reached its peak expression level after 72 h. Second, we used primer extension to validate the interaction between Yfr28 and the phosphoenolpyruvate carboxylase mRNA (ppc, PMM1575), which was ranked as its third-best predicted target (Fig. 5b). We observed distinct termination signals of ppc that did not appear in the negative control reaction with Yfr2 and ppc (Fig. 5a). These results are in full agreement with the predicted Yfr28-ppc structure complex, indicating that the synthesis of ppc cDNA ceased when the interaction of the two RNAs started (Fig. 5c). The interaction site of Yfr28 is within the last 120 nt of the ppc reading frame (Table 1). While the majority of characterized bacterial sRNAs act on the ribosome binding site (i.e., the 5′ UTR immediately upstream of the start codon), there are several sRNAs that pair deeply within the coding sequence [40,41,42]. Alternatively, or in addition, the regulation of this mRNA species by Yfr28 might also affect the 3′ adjacent gene PMM1574, encoding a GNAT family acetyltransferase. Looking at other targets, we noticed the pronounced overlap between Yfr28 and the 5′ UTR of the neighboring ftsZ gene (Table 1, Figs. S1 and S2). This 5′ UTR derives from an alternative promoter and is perfectly complementary to the first 46 nt of Yfr28 (Fig. S2). Other sRNAs with a regulatory function on ftsZ have been described in Escherichia coli [43] and Sinorhizobium meliloti [43, 44]. In Escherichia coli, it was shown that the prophage-encoded sRNA DicF inhibits cell division via direct base pairing with ftsZ mRNA to repress translation and prevent new synthesis of the protein. Robledo et al. demonstrated that Sinorhizobium meliloti sRNA EcpR1 posttranscriptionally modulates the regulation of cell cycle genes under detrimental conditions [44]. Among the top targets of Yfr28 is another cell division-related gene, the trans target minE, which encodes an ATPase-activating protein in the MinCDE system (Table 1). Interestingly, minD, encoding a membrane-bound ATPase, was in the top 40 list of EcpR1 [44]. Collectively, our data suggest that Yfr28 might play an important regulatory role affecting the cell division genes ftsZ and minE as well as ppc during nitrogen limitation. In addition, Yfr28 might be involved in the regulation of the alternative sigma factor sigD (PMM0577), which is the top five predicted target of Yfr28. The sigD homolog in Synechocystis sp. PCC6803 is sll2012, which was shown to be beneficial during nitrogen starvation, most probably because it ensures active function of genes required for maximal protection against oxidative stress and for keeping photosynthesis active [45].
Verification of Prochlorococcus NATL2A and MED4 sRNA candidates using 32P-radiolabeled specific probes. a In the well-analyzed strain MED4, two new sRNAs (Yfr28 and Yfr29) were found, the first of which is only expressed under nitrogen starvation (-N). Hybridizations with NTAL2A were performed in technical duplicates (1 and 2). The numbers given with the marker (M) are sizes in nt. b The consensus structures of HL ecotype homologs of Yfr28 and of LL ecotype homologs of Yfrs106–108 were predicted with RNAalifold [56]. The structure of the MED4-specific sRNA Yfr29 was computed with RNAfold [57].
Cultures of Prochlorococcus strain NATL2A (Yfrs106–108) or MED4 (Yfr28 and Yfr29) were exposed to various stress conditions. Total RNA extracts were separated on PAA gels, transferred to Hybond N + membranes and hybridized with the respective probes. The fold changes (normalized against the internal standard 5 S rRNA) observed under different stresses in comparison with the control hybridization are given in log2 values. The horizontal dashed gray lines indicate the border of a ≥twofold change.
a Yfr28 expression profile of Prochlorococcus MED4 samples after 0, 3, 6, 12, 24, 48, and 72 h of nitrogen starvation. The stress experiment was carried out in biological triplicates. Extracted RNA samples were separated on 7 M urea-10% PAA gels, transferred onto Hybond N+ membranes and hybridized with a Yfr28 probe. b Primer extension of 0.2 pmol of in vitro-synthesized 5′ region ppc mRNA in the absence of any synthesized sRNA or in the presence of varying amounts of Yfr28 or 160 pmol Yfr2. The vertical red line denotes the primer extension termination signals of ppc in the presence of Yfr28. The vertical cyan line indicates the interaction region predicted by intaRNA [58]. c Cofolded structures of ppc (white-circled nucleotides) and Yfr28 (royal blue-circled nucleotides). The red and cyan border lines of the nucleotides correspond to the vertical lines in (b). d Location of yfr28 (red box) in the Prochlorococcus MED4 genome. Genes of known or unknown function are indicated in blue and gray boxes, respectively, and genes encoding small RNAs are indicated in orange boxes.
One of the NATL2A sRNA candidates turned out to be an asRNA and is located opposite the IGR of atpF and atpH (encoding the b’ and delta subunits, respectively, of the ATP synthase complex), extending to the 5′ region of the atpF gene and the last 60 nt of the atpH gene (Fig. 6a). The 370 nt-long asRNA that we named as_atpF was most abundant in the 100 m sample (Fig. 6a), corresponding to the depth with the most NATL2A reads (Fig. 2). However, in northern hybridizations with total RNA from NATL2A cultures we also observed a ~70 nt long fragment (Fig. S3A) as well as longer, less distinct species of as_atpF (Fig. S3B). We further explored the functional mode of action of as_atpF by monitoring the differential expression of as_atpF, atpF, and atpH in NATL2A laboratory cultures during various stress conditions (Fig. 6b). The majority of tested stresses led to repression of the transcript levels of all three RNAs, and only modest induction of all three RNAs was observed upon HL treatment and iron starvation, suggesting that as_atpF stabilizes the mRNA of atpF and possibly also the mRNA of atpH (Fig. 6b). Furthermore, we noticed in some conditions the appearance of a short atpF mRNA fragment that is in the range of the coding sequence (462 nt) or slightly shorter, when ATP synthase might need to be stalled (especially darkness, stationary phase and cold shock, Figure S3B). This organization resembles the arrangement described for the E. coli asRNA GadY. GadY is transcribed from a promoter in the intergenic spacer between the genes gadW and gadX in antisense orientation to the 3′ end of gadX, which is an activator of the glutamate-dependent acid resistance system [46]. Upon binding of GadY, cleavage of the gadXW dicistronic mRNA is triggered [47], resulting in a more stable monocistronic gadX mRNA [48]. In MED4, similar transcriptional stress responses for atpF and atpH have been observed under darkness and HL exposure [29] and under nitrogen- [49] or iron-limiting conditions [50].
a Coverage plots of NATL2A mapped read regions in the 60 m (red), 100 m (blue) and 130 m (gray) metatranscriptome samples. The white boxes correspond to the positions of the atpF and atpH genes (encoding the b’ and delta subunit, respectively, of the ATP synthase complex). The black, orange and cyan boxes denote the positions of the probes used for the northern hybridization of as_atpF, atpF, and atpH. b Prochlorococcus NATL2A cultures were exposed to various stress conditions, and total RNA extracts were separated on PAA gels, transferred to Hybond N + membranes and hybridized with the respective probes. The fold changes (normalized against the internal standard 5 S rRNA) observed under the different stresses in comparison with the control hybridization are given in log2 values. The horizontal dashed gray line indicates the border of a ≥two fold change. The northern blots that served as basis for this analysis are shown in Fig. S3.
Conclusions and possible implications
Bacterial ncRNAs are at the heart of regulatory pathways that allow bacteria to acclimate to changes in the environment, to adjust their metabolism, to regulate the expression of virulence genes and to control many other functions [51,52,53]. Here, we present a workflow for the identification of ncRNAs that appears to be particularly applicable to bacteria of high ecological relevance that are not amenable to direct manipulation. Starting with a metatranscriptomic dataset, we focused on the important primary producer Prochlorococcus and identified several ncRNAs that are likely relevant. The sRNA Yfr28 plays a pivotal role in the coordination of primary metabolism and cell division, as indicated by its very high low-nitrogen-induced expression and identified targets, which include mRNAs encoding the cell division proteins FtsZ and MinE, phosphoenolpyruvate carboxylase, the carbon uptake proteins SbtA and B and a sigma factor. The likely effect of controlling cell division and carbon metabolism under conditions of low-nitrogen supply is physiologically reasonable. Another presented example is the asRNA of atpF, which intriguingly showed, with the exception of cold shock, the same expression responses as the mRNA to typical environmental stress conditions. The presented workflow is of particular interest for environmentally relevant microorganisms, for which experimental manipulation ability might be limiting, while abundant sequence information may be available. All scripts utilized in this workflow are freely available.
References
Michaux C, Verneuil N, Hartke A, Giard J-C. Physiological roles of small RNA molecules. Microbiology. 2014;160:1007–19.
Barquist L, Vogel J. Accelerating discovery and functional analysis of small RNAs with new technologies. Annu Rev Genet. 2015;49:367–94.
Wagner EGH, Romby P. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv Genet. 2015;90:133–208.
Georg J, Hess WR. Widespread antisense transcription in prokaryotes. Microbiol Spectr. 2018;6:191–210.
Georg J, Lalaouna D, Hou S, Lott SC, Caldelari I, Marzi S, et al. The power of cooperation: Experimental and computational approaches in the functional characterization of bacterial sRNAs. Mol Microbiol. 2019;113:603–12.
Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–27.
Biller SJ, Berube PM, Lindell D, Chisholm SW. Prochlorococcus: the structure and function of collective diversity. Nat Rev Microbiol. 2015;13:13–27.
Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73:249–99.
Moore LR, Goericke R, Chisholm SW. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar Ecol Prog Ser. 1995;116:259–75.
Lindell D, Post AF. Ultraphytoplankton succession is triggered by deep winter mixing in the Gulf of Aqaba (Eilat), Red Sea. Limnol Oceanogr. 1995;40:1130–41.
Shibl AA, Thompson LR, Ngugi DK, Stingl U. Distribution and diversity of Prochlorococcus ecotypes in the Red Sea. FEMS Microbiol Lett. 2014;356:118–26.
Biller SJ, Berube PM, Berta-Thompson JW, Kelly L, Roggensack SE, Awad L, et al. Genomes of diverse isolates of the marine cyanobacterium Prochlorococcus. Sci Data. 2014;1:140034. https://doi.org/10.1038/sdata.2014.34.
Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, Gonzalez JM, et al. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci. 2013;110:11463–8.
Axmann IM, Kensche P, Vogel J, Kohl S, Herzel H, Hess WR. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 2005;6:R73.
Steglich C, Futschik ME, Lindell D, Voss B, Chisholm SW, Hess WR. The challenge of regulation in a minimal phototroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 2008;4:e10000173.
Voigt K, Sharma CM, Mitschke J, Joke Lambrecht S, Voß B, Hess WR, et al. Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. ISME J. 2014;8:2056–68.
Steglich C, Stazic D, Lott SC, Voigt K, Greengrass E, Lindell D, et al. Dataset for metatranscriptome analysis of Prochlorococcus-rich marine picoplankton communities in the Gulf of Aqaba, Red Sea. Mar Genomics. 2015;19:5–7.
Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–64.
Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–60.
Lott SC, Voß B, Hess WR, Steglich C. CoVennTree: a new method for the comparative analysis of large datasets. Front Genet. 2015;6:43.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52.
Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, et al. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol. 2009;5:e1000502.
Otto C, Stadler PF, Hoffmann S. Lacking alignments? The next-generation sequencing mapper segemehl revisited. Bioinformatics. 2014;30:1837–43.
Lott SC, Schäfer RA, Mann M, Backofen R, Hess WR, Voß B, et al. GLASSgo–Automated and reliable detection of sRNA homologs from a single input sequence. Front Genet. 2018;9:124.
Gruber AR, Findeiß S, Washietl S, Hofacker IL, Stadler PF. RNAz 2.0: improved noncoding RNA detection. Pac Symp Biocomput. 2010;69–79.
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539.
Moore LR, Coe A, Zinser ER, Saito MA, Sullivan MB, Lindell D, et al. Culturing the marine cyanobacterium Prochlorococcus: Prochlorococcus culturing. Limnol Oceanogr Methods. 2007;5:353–62.
Pinto FL, Thapper A, Sontheim W, Lindblad P. Analysis of current and alternative phenol based RNA extraction methodologies for cyanobacteria. BMC Mol Biol. 2009;10:79.
Steglich C, Futschik M, Rector T, Steen R, Chisholm SW. Genome-wide analysis of light sensing in Prochlorococcus. J Bacteriol. 2006;188:7796–806.
Stazic D, Lindell D, Steglich C. Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res. 2011;39:4890–9.
Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–5.
Hou S, Pfreundt U, Miller D, Berman-Frank I, Hess WR. mdRNA-Seq analysis of marine microbial communities from the northern Red Sea. Sci Rep. 2016;6:35470.
Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231.
Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–7.
Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018;46:D335–D342.
Kalvari I, Nawrocki EP, Argasinska J, Quinones-Olvera N, Finn RD, Bateman A, et al. Non-coding RNA analysis using the Rfam database. Curr Protoc Bioinforma. 2018;62:e51.
Wright PR, Richter AS, Papenfort K, Mann M, Vogel J, Hess WR, et al. Comparative genomics boosts target prediction for bacterial small RNAs. Proc Natl Acad Sci USA. 2013;110:E3487–3496.
Wright PR, Georg J, Mann M, Sorescu DA, Richter AS, Lott S, et al. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014;42:W119–123.
Lambrecht SJ, Wahlig JML, Steglich C. The GntR family transcriptional regulator PMM1637 regulates the highly conserved cyanobacterial sRNA Yfr2 in marine picocyanobacteria. DNA Res. 2018;25:489–97.
Gutierrez A, Laureti L, Crussard S, Abida H, Rodríguez-Rojas A, Blázquez J, et al. β-lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat Commun. 2013;4:1610.
Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell. 2013;153:426–37.
Lalaouna D, Morissette A, Carrier M-C, Massé E. DsrA regulatory RNA represses both hns and rbsD mRNAs through distinct mechanisms in Escherichia coli: DsrA sRNA: a versatile regulator in Escherichia coli. Mol Microbiol. 2015;98:357–69.
Balasubramanian D, Ragunathan PT, Fei J, Vanderpool CK. A prophage-encoded small RNA controls metabolism and cell division in Escherichia coli. mSystems. 2016;1:e00021.
Robledo M, Frage B, Wright PR, Becker A. A stress-induced small RNA modulates alpha-rhizobial cell cycle progression. PLOS Genet. 2015;11:e1005153.
Antal T, Kurkela J, Parikainen M, Kårlund A, Hakkila K, Tyystjärvi E, et al. Roles of group 2 sigma factors in acclimation of the cyanobacterium Synechocystis sp. PCC 6803 to nitrogen deficiency. Plant Cell Physiol. 2016;57:1309–18.
Opdyke JA, Kang J-G, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;186:6698–705.
Takada A, Umitsuki G, Nagai K, Wachi M. RNase E is required for induction of the glutamate-dependent acid resistance system in Escherichia coli. Biosci Biotechnol Biochem. 2007;71:158–64.
Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol Microbiol. 2008;70:965–82.
Tolonen AC, Aach J, Lindell D, Johnson ZI, Rector T, Steen R, et al. Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability. Mol Syst Biol. 2006;2:53.
Thompson AW, Huang K, Saito MA, Chisholm SW. Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability. ISME J. 2011;5:1580–94.
Holmqvist E, Wagner EGH. Impact of bacterial sRNAs in stress responses. Biochemical Soc Trans. 2017;45:1203–12.
Carrier M-C, Lalaouna D, Massé E. Broadening the definition of bacterial small RNAs: characteristics and mechanisms of action. Annu Rev Microbiol. 2018;72:141–61.
Desgranges E, Marzi S, Moreau K, Romby P, Caldelari I. Noncoding RNA. Microbiol Spectr. 2019; 7.
Lott SC, Wolfien M, Riege K, Bagnacani A, Wolkenhauer O, Hoffmann S, et al. Customized workflow development and data modularization concepts for RNA-sequencing and metatranscriptome experiments. J Biotechnol. 2017;261:85–96.
Wright PR, Georg J. Workflow for a computational analysis of an sRNA candidate in bacteria. Bacterial Regulatory RNA. 2018. Humana Press, New York, NY, pp 3–30.
Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinforma. 2008;9:474.
Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26.
Mann M, Wright PR, Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA–RNA interactions. Nucleic Acids Res. 2017;45:W435–W439.
Acknowledgements
This work was supported by the German Science Foundation (DFG, SPP 1258) and by ASSEMBLE (Association of European Marine Biological Laboratories) Infrastructure Access Call 2 to the Interuniversity Institute for Marine Sciences (IUI), Eilat, Israel (grant agreement no: 227799), to CS, and the Federal Ministry of Education and Research (BMBF) program de.NBI-Partner grant 031L0106B to WRH. Open access funding provided by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
CS designed the study. CS, WRH and KV collected the environmental samples. CS carried out the molecular genetic and microbiological analyses. KV performed experimental sRNA validation. SJL performed temporal expression analysis of Yfr28. SCL developed the bioinformatics pipeline. CS, SCL, and WRH analyzed the data. CS, WRH and SCL drafted the manuscript with contributions from all authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lott, S.C., Voigt, K., Lambrecht, S.J. et al. A framework for the computational prediction and analysis of non-coding RNAs in microbial environmental populations and their experimental validation. ISME J 14, 1955–1965 (2020). https://doi.org/10.1038/s41396-020-0658-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-0658-7
This article is cited by
-
Soil microbial ecology through the lens of metatranscriptomics
Soil Ecology Letters (2024)