Abstract
Many of the world’s peatlands have been affected by water table drawdown and subsequent loss of organic matter. Rewetting has been proposed as a measure to restore peatland functioning and to halt carbon loss, but its effectiveness is subject to debate. An important prerequisite for peatland recovery is a return of typical microbial communities, which drive key processes. To evaluate the effect of rewetting, we investigated 13 fen peatland areas across a wide (>1500 km) longitudinal gradient in Europe, in which we compared microbial communities between drained, undrained, and rewetted sites. There was a clear difference in microbial communities between drained and undrained fens, regardless of location. Community recovery upon rewetting was substantial in the majority of sites, and predictive functional profiling suggested a concomitant recovery of biogeochemical peatland functioning. However, communities in rewetted sites were only similar to those of undrained sites when soil organic matter quality (as expressed by cellulose fractions) and quantity were still sufficiently high. We estimate that a minimum organic matter content of ca. 70% is required to enable microbial recovery. We conclude that peatland recovery after rewetting is conditional on the level of drainage-induced degradation: severely altered physicochemical peat properties may preclude complete recovery for decades.
Similar content being viewed by others
Introduction
Peatlands store vast amounts of soil carbon (C): estimates indicate that ca. 455–500 Gt C is stored in northern peatlands alone [1, 2]. This considerable C stock is increasingly destabilized by peatland degradation, which is predominantly driven by widespread water table drawdown and drought events driven by anthropogenic drainage [3] and climate change [4]. Water table drawdown causes the oxygenation of organic matter, which triggers cascading effects on microbial activity and physicochemical properties of the peat [4,5,6]. The resulting peatland degradation contributes to the emission of CO2 to the atmosphere [3], the release of dissolved organic carbon and nutrients to pore and surface water [4], and the loss of biodiversity resulting from the disappearance of specialized peatland biota [7, 8].
The primary goals of peatland restoration are to reduce carbon and nutrient losses, to restore biodiversity and peat-forming vegetation, to retain water on a landscape scale, and to eventually restore the peatland’s function as a carbon and nutrient sink. Typically, rewetting is a first crucial step in restoration attempts that aim to bring a drained peatland back to its predegradation state [3, 9, 10]. The re-establishment of waterlogged conditions is key to restore peat-forming vegetation and to attain an environment in which aerobic microbial metabolism and enzyme activity are suppressed [5], thereby creating more favorable conditions for renewed peat accumulation. Since a large share of peatlands across the globe are currently drained and therefore emitting rather than sequestering carbon [11, 12], large-scale peatland restoration by rewetting has the potential to substantially contribute to the mitigation of global warming within several decades, given that the initial increase in methane emission is outweighed by relatively fast methane turnover in the atmosphere and the peatland’s renewed carbon sink function.
However, a major uncertainty is whether rewetting alone is sufficient to fully restore drained peatlands [3, 4, 9]. Restoration hinges to a large extent on whether microbial communities, and the ecosystem functions they perform, return to the state prior to peatland degradation. This is crucial, as microbial communities are among the primary drivers of ecosystem functioning by controlling nutrient cycling and greenhouse gas emission and uptake [13].
Currently, a limited number of studies have investigated microbial community assembly after rewetting in acidic and rainwater-fed peatlands (=“bogs”) [14, 15], while even fewer studies have focused on microbial recovery in base-rich and groundwater-fed peatlands (“fen peatlands” or “fens”) [16, 17]; the dominant peatland type in the temperate zone. Among others, a recent study suggested that microbial community composition in fens is only weakly impacted by the drought-rewetting cycle [17], which would imply a high microbial resilience. However, those observations were based on a short-term experiment with a drought-rewetting cycle of only a few months, while many peatlands have been heavily drained for decades to centuries prior to rewetting. In such peatlands, multiple reasons can exist of why microbial recovery to a predegradation state may be hampered after rewetting.
First, water table drawdown alters the peat matrix through a complex interplay of cascading biogeochemical processes [4], which affect the physicochemical properties of the peat. Since microbial communities are limited by (mainly labile) carbon sources and nutrients for their metabolism and growth [4, 18], altered soil organic matter [peat] quantity and quality, an increase in bulk density, and the release of nutrients following peat oxygenation may cause long-term irreversible shifts at the microbial community level. To date, it is not well known to what extent past drought-induced shifts in physicochemical peat properties preclude microbial recovery after peatland rewetting.
Second, many undrained pristine peatlands are covered by a dense living moss carpet, i.e., primarily peat mosses (Sphagnum spp.) in bogs and members of the Amblystegiaceae family in fens. These mosses provide a continuous supply of fresh organic matter to the oxic–anoxic interface and provide an ideal habitat for many microbes, such as nitrogen-fixing and methanotrophic bacteria [15, 19, 20]. Peatland drainage generally results in a decline of moss cover and moss diversity in favor of vascular plants, while rewetting rarely leads to complete or even partial moss carpet recovery [9, 21]. Such incomplete recovery may in turn hamper recovery of the peatland bryosphere and of the microbial communities it hosts.
To investigate whether rewetting of drained fen peatlands leads to microbial recovery, we studied the vertical depth stratification of microbial communities and predicted functional gene profiles in undrained, drained and rewetted sites over a longitudinal transect in Europe. We specifically focused on prokaryotes (bacteria and archaea) as they form the dominant and highly active microbial group in peatlands [22]. We hypothesize that drainage-induced peatland degradation, including loss of organic matter and a reduction of moss cover, causes shifts in microbial community composition and in profiles of the microbes’ predicted functional genes, and that such shifts cannot simply be reversed by rewetting over the short to medium term (i.e., several years to decades) covered by our study.
Materials and methods
Study sites and sampling design
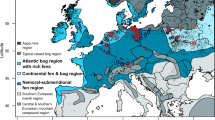
We selected a total of 39 fen sites divided over 13 regions in temperate lowland Europe, covering a wide (>1500 km) longitudinal gradient (Fig. 1 and Table S1 for metadata). The longitudinal gradient coincides with a climatological gradient from an oceanic (west, with mild winters and mild summers) to a continental (east, with cold winters and warm summers) climate. In each region, we selected three fen sites with a peat thickness of at least 0.5 m and with a different “drainage status”: one undrained site, a nearby drained site and a nearby rewetted site. Classification of sites into these three drainage categories was primarily based on our expertise in combination with knowledge on fen history from local researchers and land owners. Our expertise was based on (i) observations on the presence of drainage ditches, (ii) observations of groundwater levels at the time of sampling, and (iii) analysis of vegetation structure and composition. For undrained (i.e., near-natural) fen sites we selected sites that do not have a known history of drainage, further evidenced by a substantial cover of sedges (Carex spp.) and brown mosses (i.e., typical fen vegetation), the lack of drainage ditches, and groundwater levels near the peat surface for most of the year. Drained fen sites are characterized by obvious water table drawdown (often < 0.5 m below the surface, typically in summer), the high prevalence of drainage ditches, and the appearance of plant species typical for moist or wet grasslands. Rewetted fen sites are defined as previously drained sites in which groundwater levels have been restored to peat surface level as an outcome of local fen restoration projects or dam building by beavers. This was typically followed by the re-appearance of the aforementioned fen plant species, which also assured that rewetting had occurred at least a few years to several decades prior to our sampling campaign (successional changes in vegetation composition require several years). The exact drainage duration prior to rewetting is not known, but is in the range of many decades to centuries for all sites. Pore water pH was slightly acidic to circumneutral at all sites (6.45 ± 0.33, min = 5.6, max = 7.1; measured in the field with portable equipment, WTW Multi 340i; WTW, Weilheim, Germany).
To ascertain that the rewetted sites overall did not differ from the undrained sites in terms of current water regime, we calculated Ellenberg moisture indicator values based on vegetation composition in 2 × 2-m plots [23] (Table S1). These indicator values are considered reliable in providing a proxy for overall plot “wetness” related to soil moisture content and groundwater level [24]. We found no significant difference between moisture indicator values of rewetted (range = 7.12–9.33, mean = 8.27) and undrained (range = 7.66–8.87, mean = 8.30) sites (P < 0.98), which both were significantly higher than indicator values of the drained (range = 6.33–7.88, mean = 7.25) sites (P < 0.001, all tested by one-way ANOVA and post hoc Tukey’s HSD). In each site we selected an 8 × 4-m study plot that was covered by a homogeneous stretch of vegetation. Peat was sampled using a saw and a Russian peat corer (Eijkelkamp Soil and Water, Giesbeek, NL), with which relatively undisturbed peat cores can be collected, from the end of May through June 2017 at three depths: 0–5, 15–20, and 45–50 cm (for a total of 117 soil samples). Samples were always composed of five well-mixed subsamples (per depth). Sampling equipment was thoroughly washed with demineralized water between each use. A second set of volumetric samples (250 cm3 at depths 0–5 and 15–20 cm, and 50 cm3 at 45–50 cm) was extracted pairwise for soil bulk density calculations. All samples were individually packed in PE bags and aluminum foil, cooled immediately after collection, and deep-frozen at the end of each day.
Environmental characteristics
Soil bulk density (kg L−1) was determined by weighing oven-dried (70 °C) soil samples with known volumes. Total organic matter content was determined by loss on ignition (450 °C, 4 h).
To assess quality of the peat soil and specifically of the organic matter fraction, we determined alpha-cellulose (“cellulose” hereinafter) and acid-detergent fiber (ADF)-lignin content (“lignin” hereinafter) using the sequential ADF method [25]. In short, cetyltrimethylammonium-bromide is added to 0.5–1 g of dried and ground subsample, after which the mixture is boiled for 1 h to dissolve proteins, DNA, and most hemi-cellulose. Samples were then rinsed through a filter cup, dried at 105 °C and weighed. Next, 72% sulfuric acid (H2SO4) was added to dissolve cellulose, after which samples were again rinsed, dried, and weighed. Finally, lignin was removed from the samples by loss on ignition, and samples were again rinsed dried and weighed. Cellulose and lignin contents were expressed as fractions of total organic matter content, and are a measure of organic matter quality.
Cover of the moss layer (% of total surface) was visually estimated in a 2 × 2-m subplot within each 4 × 8-m plot.
Molecular analyses
Frozen peat samples (~600 ml each) were thoroughly homogenized through crushing with a chisel (cleaned with 70% ethanol between each sample), subsampled to ~10 g, and lyophilized. DNA was extracted from a 0.01 to 0.1 g subsample of the lyophilized peat using the PowerSoil DNA Isolation Kit (MO BIO, Carlsbad, CA, USA), following the manufacturer’s protocol, and quantified using the dsDNA Assay Kit and Qubit fluorometer (Invitrogen, Carlsbad, CA, USA).
We subjected 5 ng of DNA to a polymerase chain reaction (PCR) with the F515/R806 primer set [26] targeting an ~250 bp stretch within the V4 region of the 16S rRNA. PCR mixtures of 25 µl contained 200 nM of each primer, 1X HF buffer, 200 µM of each dNTP, and 0.25 U of Phusion High-Fidelity polymerase (Thermo Scientific, Waltham, MA, USA). PCR conditions consisted of an initial denaturation step at 98 °C for 60 s, followed by 25 cycles of: denaturation at 98 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 30 s; and an additional extension step at 72 °C for 10 min. PCR products were subsequently diluted and dual barcoded primers with Illumina adapters were added in an initial step at 98 °C for 60 s, 12 cycles at 98 °C for 10 s, 63 °C for 30 s, 72 °C for 30 s; and a final extension at 72 °C for 5 min. PCR products were run on a 1.5% agarose gel to confirm successful PCR amplification and to confirm that negative controls were empty (and in case of failure the procedure was repeated). Successful amplicons were normalized and purified using a SequalPrep Normalization Plate Kit (ThermoFisher Scientific, Waltham, MA, USA), pooled, and additionally gel extracted to completely remove primer-dimers with the Nucleospin Gel cleanup kit (Machery-Nagel, Düren, Germany). The library was then quantified through qPCR (KAPA Library Quantification Kit, Kapa Biosystems, Wilmington, MA, USA) on a BioRad CFX96 thermocycler (BioRad, Hercules, CA, USA), and sequenced on the Illumina MiSeq platform (Illumina Inc; San Diego, CA, USA) with 300 cycles for paired-end reads using V3 chemistry. Phospholipid fatty acids (PLFA’s) were quantified according to standard methodology [27,28,29], see the Supplement for a detailed description.
Taxonomic community composition
Demultiplexed paired-end reads were joined, their primers removed, filtered at 0.5 maximum expected error, clustered at 97% operational taxonomic units (OTUs), chimeric sequences discarded, and mapped to produce an OTU table using USEARCH v10 [30]. Taxonomy was assigned to representative OTUs using the SILVA 16S database (version 128) with the assign_taxonomy.py function in QIIME [31]. Non-prokaryotic 16S sequences were parsed from the OTU table leaving a total of 3,358,385 reads in the 117 samples. All samples were then rarefied to 20,000 reads per sample, leaving 24,714 unique OTUs, with the exception of 15 samples that contained less reads (between 3440 and 19,577). To allow comparison for diversity and community composition analyses, data from all samples were converted to fractions by normalization.
Prokaryote functional gene prediction and selection
OTUs (clustered at 97%) that were represented by more than 100 reads across all samples were further subjected to the PICRUSt2 program (phylogenetic investigation of communities by reconstruction of unobserved states) to predict metagenomes [32, 33].
In short, this procedure predicts the relative abundance of functional genes (expressed as Kegg Orthologs (KOs)) in a 16S OTU community from the phylogenetic conservation of these genes in all currently sequenced and assembled prokaryotic genomes. Briefly, it aligns all OTUs to a reference tree after which it predicts the presence and abundance of genes from their phylogenetic placement relative to the fully sequenced genomes, accounting for 16S copy numbers. For our dataset, this resulted in a total of 7029 KOs (=“Complete KO dataset”). In addition to this complete dataset, we also selected a small set of functional genes that we perceived as relevant for fen functioning, with a focus on nitrogen (N), methane (CH4), and sulfur (S) cycling. To do this, we performed an automated query of all KOs that contained either of the words ‘ammon’, ‘nitr’, ‘meth’, ‘sulfa’, and ‘sulfi’ in the function description, after which two researchers (WJE and EV) scored (1–10 scale) the remaining KOs for perceived relevance to nutrient and methane cycling. Relevance was assessed by looking up the individual KOs in the KEGG-database (Accessed December 2018, https://www.genome.jp/kegg), as well as by a query in Web of Science to see if the KO matches with a relevant (=N, S, or CH4 related) process. Both scores were positively correlated (r = 0.7, P < 0.001), and only KOs with a cumulative score of at least 17/20 were included. For closely related KOs (e.g., methyl-coenzyme M reductase alpha and beta subunits (mcrA and mcrB) [EC:2.8.4.1]) we only randomly selected one KO, resulting in a final selection of 24 KOs (=“Reduced KO dataset”). Relative functional gene abundance per sample was calculated by dividing the number of predicted genes per sample by the total number of reads per sample.
As a check, we verified the accuracy of PICRUSt2 predictions using one microbial test group, i.e., methanogens, with qPCR on a set of 72 randomly selected peat samples using the same 16S primers as above (but without multiplexing adapters) and primers targeting the methyl-coenzyme-M reductase (mcrA) gene (responsible for methanogenesis) [34]. Prior to this qPCR all DNA isolations were additionally cleaned using one-step PCR inhibitor removal kit (Zymo research, Irvine, CA, USA) and re-quantified using Qubit. qPCR was performed in duplicate reactions using KAPA SYBR fast (Roche, Basel, Switzerland) with the following cycling conditions: 95 °C for 180s followed by 40 cycles at 95 °C for 3 s, 57 °C for 20 s, and 72 °C for 12 s. For standards, diluted cleaned-up PCR products were used and it was ascertained that accuracy was high through a linear model of log-transformed concentrations on Cq’s (R2 > 0.993). Copy numbers of genes were estimated through Qubit measures of standards assuming a length in bp’s of 292 for 16S and 478 for mcrA. Finally, we divided mcrA copy numbers by total 16S copy numbers to obtain relative mcrA abundances (per sample), which we then compared with relative mcrA abundances as predicted by PICRUSt2.
Statistical analyses
All analyses were performed in R 3.5.1 and significance for statistical tests was accepted at P < 0.05.
For data visualization, we performed nonmetric multidimensional scaling (k = 2) based on Bray–Curtis dissimilarity indices for the normalized (1) sample*OTU taxonomy matrix (97% clustering) and (2) the sample*KO functional gene matrices (Repeated for both the “Complete” and the “Reduced” KO dataset). Differences in community composition and predicted functional genes across depth and fen drainage status, and the interaction between these two factors, were tested with permutational analysis of variance (permanova) with 999 iterations, using the function ‘adonis’ in which we included plot location as a blocking factor (‘strata’) to account for within-plot dependence of the three depths. Environmental variables that were significantly correlated with microbial community composition were visualized as vectors onto the ordination (‘envfit’ function with 999 iterations). Next, we calculated Shannon diversity indices within each sample as a measure for prokaryote alpha diversity.
In addition to these main analyses at the 97% OTU level, we also aggregated data at the phylum level for the 15 phyla with highest relative abundances (the remaining phyla were grouped in a rest group, which had individual across-sample mean relative abundances <0.6%).
Next, linear-mixed effect models were used to test for the effects of fen drainage status (factor: undrained–rewetted–drained) and depth (as factor) on relative abundance of each phylum, Shannon diversity index and prokaryote PLFA concentration. Plot location was included in the model as a random factor to account for within-plot dependence of the three depth classes. If the interaction term in the models were nonsignificant, we re-ran the tests including main effects only. PLFA concentrations first had to be log10 transformed to conform to normality of the model residuals.
For the following analyses we focused on the top (0–5 cm) peat layer only as this is (i) the biologically most active layer and (ii) primarily and most heavily affected by water table drawdown. We compared the predicted relative abundance of selected genes (“Reduced KO dataset”) that contribute to dominant biogeochemical processes in drained and rewetted fens to their relative abundances in undrained fens (=control group), and we calculated the standardized mean differences (SMD-H) of each of the selected KOs between the undrained–rewetted (U–R) fens and the undrained–drained (U–D) fens.
Finally, for all individual samples collected in the top peat layer, we calculated microbial community dissimilarities in relation to average taxa abundances in undrained fens, which was done by averaging all Bray–Curtis distances from a sample to all samples collected in the top layer of undrained fens (based on the sample*OTU taxonomy matrix (with 97% clustering)). We then correlated (Pearson’s r) these mean microbial community dissimilarities to the environmental variables measured in rewetted fens and in drained fens to investigate which environmental factors explain whether the microbial community resembles undrained fens after rewetting.
Results
Prokaryote community composition, biomass, and diversity
We used high-throughput amplicon sequencing with 97% OTU clustering to determine prokaryote community composition in fen sites that differed in drainage status (drained, rewetted, and undrained). Community composition was primarily determined by depth (Permanova pseudo-F = 20.34, R2 = 0.248, P < 0.001) and to a lesser extent by drainage status (Permanova pseudo-F = 4.45, R2 = 0.055, P < 0.001), with a significant interaction effect between depth and fen drainage status (Permanova pseudo-F = 1.51, R2 = 0.037, P = 0.031, Fig. 2). Differences in microbial community composition that were related to fen drainage status were most pronounced in the top (0–5 cm) soil (R2 top layer only = 0.132, R2 mid layer only = 0.121, R2 deepest layer only = 0.116). Here, microbial community composition differed between drained and undrained fens (Permanova pseudo-F = 4.94, R2 = 0.170, P < 0.001) and to a lesser extent also between drained and rewetted fens (Permanova pseudo-F = 2.23, R2 = 0.085, P = 0.013), but we found no significant differences in overall community composition between undrained and rewetted fens (Permanova pseudo-F = 1.27, R2 = 0.052, P = 0.168).
The analysis is based on Bray–Curtis dissimilarity indices for the normalized (1) sample*OTU taxonomy matrix (97% clustering after 16S rRNA sequencing). Variables (vectors) that are significantly correlated with microbial community composition were rescaled and moved from the center of the ordination to the top left corner.
Furthermore, there was a distinct vertical depth stratification in most phyla: Actinobacteria, Acidobacteria, and Bacteroidetes had a higher relative abundance near the soil surface, whereas Bathyarchaeota, Chloroflexi, Ignavibacteriae, Euryarchaeota, and Nitrospirae were relatively more abundant in one of the deeper layers (Fig. S1, Table S2). For several phyla there was an interaction effect between depth and fen drainage status: Aminicenantes (overall increase with depth) and Proteobacteria (overall decrease with depth) showed a much more pronounced vertical stratification in undrained fens compared with drained fens, whereas the opposite pattern was visible for Spirochaetae (overall increase with depth) (Fig. S1, Table S2). Acidobacteria, Gemmatimonadetes, and Nitrospirae were relatively more abundant in drained fens than in undrained fens, whereas Chloroflexi were more abundant in undrained fens. Shannon diversity index H (based on 97% OTU clustering) was also determined by an interaction effect between fen drainage status and depth (F = 3.9, P = 0.006, Fig. 3a). Highest prokaryote diversity was found in the topsoil of undrained fens, which also exhibited a more distinct depth stratification (Fig. 3a). Diversity in drained fens was generally lower and showed no significant decrease with depth. Total prokaryote biomass as estimated by PLFA concentration decreased with depth (F = 296.6, P < 0.001) and also differed between fens of different drainage status (F = 3.88, P = 0.030), with a higher concentration in drained fens (Fig. 3b).
Predicted functional profiles and potential biogeochemical shifts
The composition of predicted prokaryote functional genes was mainly determined by depth (Complete dataset and reduced dataset respectively: Permanova pseudo-F = 41.47 and 99.39, R2 = 0.404 and 0.606, P < 0.001, Fig. S2) and by fen drainage status (Complete dataset and reduced dataset, respectively: Permanova pseudo-F = 5.30 and 8.60, R2 = 0.0516, and 0.0525, P < 0.001, Fig. S2). In the top layer, the relative abundances of the predicted genes related to methanogenesis, methane oxidation, N fixation, nitrate respiration and dissimilatory sulfate reduction were on average lower in drained fens than in undrained fens (Fig. 4). In contrast, genes related to denitrification and nitrate reduction (both assimilatory and dissimilatory) and assimilatory sulfate reduction tended to be relatively more abundant in drained fens than in undrained fens (Fig. 4). Rewetted fens occupied an intermediate position between drained and undrained fens and 95% confidence intervals of the abundance estimates overlapped in most cases with the baseline of average values for undrained fens (Fig. 4), suggesting a high overall similarity between relative functional gene abundance in rewetted and undrained fens. For the mcrA gene, PICRUSt2 predictions of relative mcrA abundance correlated with estimations based on qPCR (R2 = 0.59, df = 70, P < 0.001, Fig. S3).
(Top panel): Standardized differences of the means (±95% CI) of relative abundances of predicted functional genes in the top layer of drained (dots) and rewetted (triangles) fens versus undrained fens (=baseline control, dashed line). Values below and above the baseline respectively indicate a lower and higher relative gene abundance in drained/rewetted fens compared with undrained fens. (Bottom panel): Relative abundances (±95% CI) of predicted functional genes in undrained fens.
Environmental controls on microbial community assembly
Microbial community compositions in the top layer of rewetted fens were more dissimilar to those of undrained fens with decreasing organic matter content (P = 0.005, df = 11, Fig. 5a) and with increasing bulk density (P = 0.006, df = 11, Fig. 5b). Within the topsoil of undrained reference peatlands, the maximum microbial community dissimilarity equaled 0.612 (excluding one outlier, vertical boxplots in Fig. 5). By using this boundary value in the modeled relationship between microbial community dissimilarities versus organic matter contents in rewetted fens (i.e., regression line in Fig. 5a, with equation y = −0.0022*x + 0.7722), we estimated that a minimum topsoil organic matter content of 72.8% (i.e., ca. 70%) would be required for a rewetted fen to be within the microbial community dissimilarity range of undrained fens.
Environmental variables include: a soil organic matter content (%), b soil bulk density (kg L−1), c cellulose fraction of organic matter, d lignin fraction of organic matter, e longitude of the study site, and f moss cover (%). Microbial community dissimilarities are based on Bray–Curtis dissimilarity results (after 16S rRNA sequencing and 97% OTU clustering) from each individual (drained or rewetted) sample of the top peat layer (0–5 cm) to all the samples collected in the top layer of undrained fens: the larger the dissimilarity, the more the microbial community deviates from communities in undrained fens. The horizontal boxplots represent the range of the environmental values in undrained fens, the vertical boxplots represent the range of microbial community dissimilarities within undrained fens only. Pearson’s correlation coefficients (r) are given in each plot; significance is indicated with **P < 0.01.
Furthermore, microbial community dissimilarities increased with a decreasing cellulose fraction of organic matter (P = 0.003, df = 11, Fig. 5c), while there was no correlation with lignin fractions (P = 0.442, df = 11, Fig. 5d).
We found no significant correlation between microbial community dissimilarities in rewetted fens (compared with undrained fens) with the climatological/longitudinal gradient (P = 0.333, df = 11, Fig. 5e) or with moss cover (P = 0.750, df = 11, Fig. 5f). For drained fens, none of the environmental variables correlated with microbial community dissimilarity from undrained fens (Fig. 5).
Discussion
We studied microbial communities in undrained, drained, and rewetted fens across a climatological gradient in temperate Europe to investigate whether rewetting of drained peatlands leads to recovery of the communities and their predicted functional profiles. We found that the microbiome can often recover from drainage through the process of rewetting, as microbial communities in rewetted fens were usually within the natural range of variation of undrained fens. This response was independent of location along the gradient. However, in line with our hypothesis, highest recovery was observed in those rewetted fens where the physicochemical properties of the peat soil, as expressed by organic matter quantity and quality, had not been severely altered.
Depth stratification
Depth was the primary determinant of microbial community composition and functional profiles, in line with most previous studies [35,36,37]. In other words, microbial communities at equal depths in different fens were generally more similar than were microbial communities at different depths within the same fen. Also in line with the majority of studies [37,38,39,40], prokaryote diversity was highest in the top peat layer and decreased with depth. A similar trend was observed for prokaryote biomass, which decreased approximately an order of magnitude across the depth profile. The explanatory effect of fen drainage status on microbial communities was most pronounced in the topsoil and decreased with increasing depth. This result was expected: (1) drainage-induced peat oxidation and degradation starts in the top peat layer and then gradually progresses into deeper layers, and (2) deeper layers are usually anoxic regardless of drainage status. The dominant effect of depth on microbial communities is likely imposed by increasing energetic constraints caused by a vertical stratification in the availability of O2 and of alternative electron acceptors, carbon [peat/substrate] quality, quantity and input rate, and possible dispersal limitations due to a decreased hydraulic conductivity and peat compaction [37]. Therefore, microbes at greater depths require specific specializations to thrive in such energetically constraining and oxygen-depleted environments [36, 37, 41]. In support, we indeed found a higher relative abundance of archaeal phyla such as Bathyarchaeota or Euryarchaeota with increasing depth (Fig. S1), which are known to thrive in anoxic organic habitats.
Environmental controls on microbial recovery in rewetted fens
It has been suggested that microbial communities are quite resilient to shifts in substrate quality brought about by land use change [42], or to the drainage-rewetting cycle [17]. In our study, microbial communities of rewetted fens were largely similar to communities of undrained fens, also suggesting substantial recovery by rewetting. This pattern appeared independent of location along the climatological (=oceanic-continental) gradient. However, there was also a high variability in recovery among rewetted fens. Subsequent correlative analyses revealed that a higher microbial community dissimilarity was observed in rewetted fens in which soil organic matter contents were relatively low, i.e., with an estimated organic matter content less than ca. 70%. This threshold estimate should be seen as indicative: the negative linear relationship between organic matter content and microbial community dissimilarity suggests a continuum of decreasing rewetting success with decreasing organic matter content, without indicating a tipping point.
We believe that the observed shifts in microbial community composition in rewetted fens should be a direct consequence of drainage-induced shifts in physicochemical properties of the peat matrix, as the latter are not quickly reversed by rewetting [4, 21]. Drainage and concomitant peat oxidation result in rapid loss of organic matter and subsequent alterations of carbon sources, nutrient release, and soil compaction [4, 43, 44], while undrained peatlands are characterized by high-organic matter contents and low-bulk densities as also confirmed by our study (horizontal boxplots in Fig. 5a, b).
In support, our data also showed that drained and rewetted fens on average had a lower cellulose fraction than undrained fens, which suggests that not only quantity but also quality of organic matter was altered. Cellulose is the most abundant carbohydrate in plants and is easier to decompose than lignin, especially in anoxic environments [45], and its hydrolysis is an important source of energy and carbon for peatland microbes.
In addition to the aforementioned results, it is possible that microbial recovery in rewetted fens may be co-determined by other factors such as time since rewetting or overall rewetting success, which differs between the rewetted study sites (Table S1). Although the lack of accurate data precludes detailed analyses including these two variables, a classification of the rewetted fens in our study in short (<10 years, four fens), medium (10–25 years, five fens), and longer (>25 years, four fens) time since rewetting suggests that there were no significant differences between these groups in (i) fen organic matter contents or (ii) microbial community dissimilarities to undrained fens (Fig. S4; Spearman’s correlation test, P > 0.05 for both variables). This may be because the fens in our dataset were rewetted only between a few years and several decades ago, which is probably a too short time span to significantly reverse drainage-induced physicochemical shifts: even under good conditions, fen peat accretion rates are estimated to be as low as 0.35–0.89 mm year−1 [46, 47]. Furthermore, Ellenberg moisture indicator values of the rewetted fens, which can be used as a proxy for overall site wetness and thus for overall “rewetting success”, also did not correlate significantly with fen organic matter contents (Spearman’s rho = −0.03, P = 0.906) or with microbial community dissimilarities to undrained fens (Spearman’s rho = −0.33, P = 0.27).
Finally, we also hypothesized that a drainage-induced reduction of moss cover may cause shifts in microbial community composition. Recovery of a dense moss carpet may be an additional prerequisite for microbiome recovery in rewetted peatlands, as it characterizes the vegetation of undrained peatlands (horizontal boxplot in Fig. 5f). Although this statement probably holds for specific groups such as methanotrophs [15, 20, 48], we found no significant correlation between moss cover in rewetted sites and overall microbial community dissimilarity to undrained sites. It is known from bogs that especially Sphagnum cover determines whether a rewetted bog resembles pristine bogs over a similar time-scale as studied here [15], however we see no such trends in the cover of brown mosses in our fens.
In the drained fen sites, none of the predefined environmental variables correlated significantly with microbial community dissimilarity to undrained sites. This indicates that other (unmeasured) environmental variables determine prokaryote community composition in the drained sites, among which high oxygen availability and high redox-potential are plausible candidates.
Taken together, our results suggest that drainage-induced and short-term irreversible changes in quantity and quality of soil carbon, as well as changes in quality or input rates of fresh organic compounds [49, 50], may constrain microbiome recovery upon rewetting. It is plausible that the rewetted sites with a more intact physicochemical peat matrix may have only been subject to relatively shorter and milder drainage histories, which make them more easily restorable to predegradation conditions.
Degradation-induced shifts in functional profiles of peatland microorganisms
We observed clear taxonomic shifts from undrained to drained fen sites: drained sites contained a higher proportion of the phyla Acidobacteria, Gemmatimonadetes, and Nitrospirae, whereas undrained sites contained relatively more Chloroflexi. Since most prokaryote phyla contain members that occur in a wide range of contrasting environments, shifts in taxonomy on the phylum level are not easily translated to shifts in fen functioning.
Here, predictive functional profiling of taxa provides more insight in potential shifts in ecosystem functioning. Functional profiling indeed revealed a strong functional distinction between drained and undrained sites. Drained sites were, for example, characterized by a lower relative abundance of genes involved in methanogenesis, methane oxidation, N fixation, nitrate respiration, and dissimilatory sulfate reduction in the top layer, while genes related to denitrification and nitrate reduction (both assimilatory and dissimilatory) and assimilatory sulfate reduction were relatively more abundant in drained sites. However, shifts in relative gene abundances still need to be interpreted with caution, mainly because (i) shifts are relative and do not represent shifts in absolute abundance and (ii) the function of certain genes is not always obvious. Nonetheless, shifts in some well-known genes are easily interpretable. For instance, the observed drop in methane-related genes (methanogenesis (e.g., mcrA) and methanotrophy (e.g., pmoA)) after drainage was expected: methanogenesis relies on waterlogged and anoxic conditions, whereas microbe-mediated methane oxidation relies on a continuous supply of CH4 on the oxic–anoxic interface [10]. Rewetted sites occupied an intermediate position between drained and undrained sites in terms of relative functional gene abundance, but the 95% CIs for all but three genes (narP, nifH, and cysH genes) overlapped with the undrained baseline (Fig. 4). These results suggest a substantial prokaryote functional recovery after rewetting, in line with other results presented in this study.
Conclusion
We provide empirical evidence that a substantial recovery of microbial communities and their functional profiles can be accomplished by rewetting drained fen peatlands, indicating high levels of microbial resilience. However, drainage legacies, such as the loss of organic matter quantity and quality, which are not easily reversed, may preclude complete recovery on a decadal time span. We stress that additional drainage of hitherto undrained peatlands should therefore be avoided, and the immediate rewetting of recently or mildly drained peatlands is of pivotal importance to quickly restore peatland functioning. Conversely, rewetting of long-term or severely drained peatlands may result in a substantial reduction of greenhouse gas emissions [51], but it may not result in full microbiome recovery within a reasonable time span.
Data availability
Sequences are available in NCBI SRA under project number PRJNA595701.
References
Gorham E. Northern peatlands: role in the carbon cycle and probable responses to climatic warming. Ecol Appl. 1991;1:182–95.
Yu Z. Northern peatland carbon stocks and dynamics: a review. Biogeosciences. 2012;9:4071–85.
Knox SH, Sturtevant C, Matthes JH, Koteen L, Verfaillie J, Baldocchi D. Agricultural peatland restoration: effects of land-use change on greenhouse gas (CO2 and CH4) fluxes in the Sacramento-San Joaquin Delta. Glob Change Biol. 2015;21:750–65.
Fenner N, Freeman C. Drought-induced carbon loss in peatlands. Nat Geosci. 2011;4:895–900.
Freeman C, Ostle N, Kang H. An enzymic ‘latch’ on a global carbon store - a shortage of oxygen locks up carbon in peatlands by restraining a single enzyme. Nature. 2001;409:149.
Kwon MJ, Haraguchi A, Kang H. Long-term water regime differentiates changes in decomposition and microbial properties in tropical peat soils exposed to the short-term drought. Soil Biol Biochem. 2013;60:33–44.
Laine J, Vasander H, Sallantaus T. Ecological effects of peatland drainage for forestry. Environ Rev. 1995;3:286–303.
Minayeva TY, Bragg O, Sirin A. Towards ecosystem-based restoration of peatland biodiversity. Mires Peat. 2017;19:1–36.
Mälson K, Backéus I, Rydin H. Long-term effects of drainage and initial effects of hydrological restoration on rich fen vegetation. Appl Veg Sci. 2007;11:99–106.
Juottonen H, Hynninen A, Nieminen M, Tuomivirta TT, Tuittila ES, Nousiainen H, et al. Methane-cycling microbial communities and methane emission in natural and restored peatlands. Appl Environ Micro. 2012;78:6386–9.
Lamers LP, Vile MA, Grootjans AP, Acreman MC, van Diggelen R, Evans MG, et al. Ecological restoration of rich fens in Europe and North America: from trial and error to an evidence‐based approach. Biol Rev. 2015;90:182–203.
Joosten H. The Global Peatland CO2 Picture: peatland status and drainage related emissions in all countries of the world. Ede: Wetlands International; 2009.
Bardgett RD, Van Der Putten WH. Belowground biodiversity and ecosystem functioning. Nature. 2014;515:505.
Andersen R, Francez A-J, Rochefort L. The physicochemical and microbiological status of a restored bog in Québec: Identification of relevant criteria to monitor success. Soil Biol Biochem. 2006;38:1375–87.
Putkinen A, Tuittila E-S, Siljanen HM, Bodrossy L, Fritze H. Recovery of methane turnover and the associated microbial communities in restored cutover peatlands is strongly linked with increasing Sphagnum abundance. Soil Biol Biochem. 2018;116:110–9.
Wen X, Unger V, Jurasinski G, Koebsch F, Horn F, Rehder G, et al. Predominance of methanogens over methanotrophs in rewetted fens characterized by high methane emissions. Biogeosciences. 2018;15:6519–36.
Potter C, Freeman C, Golyshin PN, Ackermann G, Fenner N, McDonald JE, et al. Subtle shifts in microbial communities occur alongside the release of carbon induced by drought and rewetting in contrasting peatland ecosystems. Sci Rep. 2017;7:11314.
Sinsabaugh RL, Hill BH, Shah JJF. Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature. 2009;462:795–U117.
Larmola T, Leppänen SM, Tuittila E-S, Aarva M, Merilä P, Fritze H, et al. Methanotrophy induces nitrogen fixation during peatland development. PNAS. 2014;111:734–9.
Liebner S, Zeyer J, Wagner D, Schubert C, Pfeiffer EM, Knoblauch C. Methane oxidation associated with submerged brown mosses reduces methane emissions from Siberian polygonal tundra. J Ecol. 2011;99:914–22.
Aggenbach CJ, Backx H, Emsens WJ, Grootjans AP, Lamers LP, Smolders AJ, et al. Do high iron concentrations in rewetted rich fens hamper restoration. Preslia. 2013;85:405–20.
Myers B, Webster KL, Mclaughlin JW, Basiliko N. Microbial activity across a boreal peatland nutrient gradient: the role of fungi and bacteria. Wetl Ecol Manag. 2012;20:77–88.
Ellenberg H, Leuschner C. Vegetation Mitteleuropas mit den Alpen: in ökologischer, dynamischer und historischer Sicht. Stuttgart: Ulmer Verlag; 2010. Vol. 8104.
Schaffers AP, Sýkora KV. Reliability of Ellenberg indicator values for moisture, nitrogen and soil reaction: a comparison with field measurements. J Veg Sci. 2000;11:225–44.
Rowland A, Roberts J. Lignin and cellulose fractionation in decomposition studies using acid‐detergent fibre methods. Commun Soil Sci Plan. 1994;25:269–77.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS. 2011;108:4516–22.
Frostegard A, Baath E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fert Soils. 1996;22:59–65.
Oravecz O, Elhottová D, Krištůfek V, Šustr V, Frouz J, Tříska J, et al. Application of ARDRA and PLFA analysis in characterizing the bacterial communities of the food, gut and excrement of saprophagous larvae ofPenthetria holosericea (Diptera: Bibionidae): a pilot study. Folia microbiologica. 2004;49:83.
Šnajdr J, Valášková V, Merhautová V, Cajthaml T, Baldrian P. Activity and spatial distribution of lignocellulose-degrading enzymes during forest soil colonization by saprotrophic basidiomycetes. Enzym Micro Tech. 2008;43:186–92.
Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6.
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814.
Douglas GM, Maffei VJ, Zaneveld J, Yurgel SN, Brown JR, Taylor CM et al. PICRUSt2: an improved and extensible approach for metagenome inference. 2019. https://www.biorxiv.org/content/10.1101/672295v1.
Luton PE, Wayne JM, Sharp RJ, Riley PW. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology. 2002;148:3521–30.
Galand PE, Saarnio S, Fritze H, Yrjälä K. Depth related diversity of methanogen Archaea in Finnish oligotrophic fen. FEMS Microbiol Ecol. 2002;42:441–9.
Lüdemann H, Arth I, Liesack W. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl Environ Microbiol. 2000;66:754–62.
Andersen R, Chapman SJ, Artz RRE. Microbial communities in natural and disturbed peatlands: a review. Soil Biol Biochem. 2013;57:979–94.
Morales SE, Mouser PJ, Ward N, Hudman SP, Gotelli NJ, Ross DS, et al. Comparison of bacterial communities in New England Sphagnum bogs using terminal restriction fragment length polymorphism (T-RFLP). Micro Ecol. 2006;52:34–44.
Eilers KG, Debenport S, Anderson S, Fierer N. Digging deeper to find unique microbial communities: the strong effect of depth on the structure of bacterial and archaeal communities in soil. Soil Biol Biochem. 2012;50:58–65.
Jackson CR, Liew KC, Yule CM. Structural and functional changes with depth in microbial communities in a tropical malaysian peat swamp forest. Micro Ecol. 2009;57:402–12.
Brune A, Frenzel P, Cypionka H. Life at the oxic–anoxic interface: microbial activities and adaptations. FEMS Microbiol Rev. 2000;24:691–710.
Basiliko N, Henry K, Gupta V, Moore T, Driscoll B, Dunfield P. Controls on bacterial and archaeal community structure and greenhouse gas production in natural, mined, and restored Canadian peatlands. Front Microbiol. 2013;4:215.
Hooijer A, Page S, Jauhiainen J, Lee WA, Lu XX, Idris A, et al. Subsidence and carbon loss in drained tropical peatlands. Biogeosciences. 2012;9:1053–71.
Minkkinen K, Laine J. Effect of forest drainage on the peat bulk density of pine mires in Finland. Can J For Res. 1998;28:178–86.
Pérez J, Munoz-Dorado J, De la Rubia T, Martinez J. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview. Int Microbiol. 2002;5:53–63.
Borren W, Bleuten W, Lapshina ED. Holocene peat and carbon accumulation rates in the southern taiga of western Siberia. Quat Res. 2004;61:42–51.
Succow M, Joosten H. landschaftsökologische Moorkunde. Stuttgart: E. Schweizerbart’sche Verlagsbuchhandlung; 2012.
Raghoebarsing AA, Smolders AJ, Schmid MC, Rijpstra WIC, Wolters-Arts M, Derksen J, et al. Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature. 2005;436:1153.
Eilers KG, Lauber CL, Knight R, Fierer N. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol Biochem. 2010;42:896–903.
Cleveland CC, Nemergut DR, Schmidt SK, Townsend AR. Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition. Biogeochemistry. 2007;82:229–40.
Hiraishi T, Krug T, Tanabe K, Srivastava N, Baasansuren J, Fukuda M et al. 2013 supplement to the 2006 IPCC guidelines for national greenhouse gas inventories: Wetlands. Switzerland: IPCC; 2014.
Acknowledgements
We are grateful to Johan de Gruyter for help with operating the sequencer, and we thank Tom van der Spiet, Rebecca White, and Adrita Ballal for their assistance in the lab. We thank Natuurpunt, Staatsbosbeheer, Natagora, Stiftung Naturschutz Schleswig-Holstein, and the districts of Vorpommern-Rügen and Vorpommern-Greifswald for sampling permits, and greatly thank Filip Meysman, Tim Urich, Richard Bardgett, and three anonymous reviewers for critical reading of and input on the manuscript. Finally our gratitude to all field helpers and all members of the REPEAT team. This research was financed by BiodivERsA/BELSPO (BR/175/A1), VBNE/OBN (OBN-2016-80-NZ), and FWO (1214520N).
Author information
Authors and Affiliations
Contributions
W-JE, EV, RvD, CJSA, AK, WK, LK, ES, HS, FT, and MW designed the research. RvD, CJSA, TC, JF, LK, YL, ES, FT, and JV collected the data. W-JE and EV statistically analyzed the data. W-JE led the writing of the manuscript and all authors contributed to drafts and gave approval for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Emsens, WJ., van Diggelen, R., Aggenbach, C.J.S. et al. Recovery of fen peatland microbiomes and predicted functional profiles after rewetting. ISME J 14, 1701–1712 (2020). https://doi.org/10.1038/s41396-020-0639-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-0639-x
This article is cited by
-
Unraveling microbial processes involved in carbon and nitrogen cycling and greenhouse gas emissions in rewetted peatlands by molecular biology
Biogeochemistry (2024)
-
Status of Drug Discovery in Wetlands Through a Lens of Bioprospecting for New Antimicrobials Being Produced by Microorganisms
Current Clinical Microbiology Reports (2023)
-
Decomposition of lignin and carbohydrates in a rewetted peatland: a comparative analysis of surface water and anaerobic soil layers
Biogeochemistry (2023)
-
An examination of the influence of drained peatlands on regional stream water chemistry
Hydrobiologia (2023)
-
Shifts in the Rhizosphere and Endosphere Colonizing Bacterial Communities Under Drought and Salinity Stress as Affected by a Biofertilizer Consortium
Microbial Ecology (2022)