Abstract
In the context of infection, Pseudomonas aeruginosa and Staphylococcus aureus are frequently co-isolated, particularly in cystic fibrosis (CF) patients. Within lungs, the two pathogens exhibit a range of competitive and coexisting interactions. In the present study, we explored the impact of S. aureus on the physiology of P. aeruginosa in the context of coexistence. Transcriptomic analyses showed that S. aureus significantly and specifically affects the expression of numerous genes involved in P. aeruginosa carbon and amino acid metabolism. In particular, 65% of the strains presented considerable overexpression of the genes involved in the acetoin catabolic (aco) pathway. We demonstrated that acetoin is (i) produced by clinical S. aureus strains, (ii) detected in sputa from CF patients and (iii) involved in P. aeruginosa’s aco system induction. Furthermore, acetoin is catabolized by P. aeruginosa, a metabolic process that improves the survival of both pathogens by providing a new carbon source for P. aeruginosa and avoiding the toxic accumulation of acetoin on S. aureus. Due to its beneficial effects on both bacteria, acetoin catabolism could testify to the establishment of trophic cooperation between S. aureus and P. aeruginosa in the CF lung environment, thus promoting their persistence.
Similar content being viewed by others
Introduction
Infectious sites constitute rich microbial ecosystems shared by a large diversity of microorganisms, including the native microbiota and pathogens. Lungs of cystic fibrosis (CF) patients are a well-known example of this microbial richness as they gather more than 60 genera of bacteria [1, 2]. This density of microorganisms promotes their interactions, through which they model their biological activities and their environment [3, 4]. However, microbial interactions are dynamic and range from antagonism to cooperation according to species and environmental conditions [5]. For instance, the opportunistic pathogen Pseudomonas aeruginosa is well known for its competitiveness in various ecosystems due to many quorum-sensing-mediated factors, such as phenazines, rhamnolipids and the type 6 secretion system [6]. P. aeruginosa thus can alter the growth, biofilm formation and respiration of yeasts, fungi and Gram-negative and Gram-positive bacteria [6]. Among them, Staphylococcus aureus is particularly sensitive to P. aeruginosa virulence factors as they can directly lyse staphylococci [7]. This competitive interaction between P. aeruginosa and S. aureus is observed for both environmental and clinical strains as both pathogens are frequently co-isolated from wounds and CF lung samples [7, 8]. Until recently, the anti-staphylococcal behaviour of P. aeruginosa was the only expression observed between the two species and was thus extensively described [7]. In the context of CF lung infections, this competitive interaction is highlighted by the decreased prevalence of S. aureus as P. aeruginosa colonizes lungs during adolescence [9]. However, Baldan et al. [10] first noted that a non-competitive state between P. aeruginosa and S. aureus could establish during the development of CF chronic infections, calling into question the antagonistic model between the two pathogens.
The establishment of this particular non-competitive state between P. aeruginosa and S. aureus seems to be linked to the adaptation of P. aeruginosa to the pulmonary ecosystem. In fact, selective pressures present in the CF lung environment, such as the host immune system and antibiotic treatments, drive P. aeruginosa isolates towards a state of low virulence and high resistance [11,12,13,14]. Major virulence factors involved in quorum sensing and motility are often mutated in P. aeruginosa chronic infection isolates, inducing a decrease in the production of anti-staphylococcal factors followed by non-competitive interaction [15]. In addition, the rewiring of metabolism networks and the decrease of its catabolic repertoire also accompany P. aeruginosa’s adaptation to the CF environment. This trophic specialization commonly leads to the slower growth of chronic isolates and thus less competitive behaviour regarding shared resources [16]. Several independent studies observed this state of coexistence between S. aureus and P. aeruginosa isolated from chronic infections [10, 15, 17, 18]. Briaud et al. [18, 19] recently demonstrated that this interaction pattern appears to be more frequent than expected. Indeed, among the quarter of CF patients co-infected by both pathogens, 65% were infected by a coexisting S. aureus–P. aeruginosa pair. Recent studies have shown that coexistence between P. aeruginosa and S. aureus could promote their persistence throughout the establishment of cooperative interaction. In these conditions, coexisting bacteria demonstrated increased tolerance to antibiotics: to tobramycin and tetracycline for S. aureus and to gentamicin for P. aeruginosa. This appeared to be related to the induction of small colony variants [15, 17, 18, 20]. However, the effects of coexistence on general bacterial physiology, and not only virulence-associated traits, have not yet been explored. Therefore, despite its significance in infectious ecosystems, coexistence between P. aeruginosa and S. aureus remains poorly understood.
Using global and targeted transcriptomic approaches, we evaluated the impact of the presence of S. aureus on the gene expression of P. aeruginosa using a set of clinical pairs of strains isolated from CF co-infected patients. Coexistence with S. aureus induced the overexpression of many genes involved in the utilization of alternative carbon sources in P. aeruginosa, such as amino acids and acetoin. Acetoin was shown to be produced by clinical S. aureus isolates in vitro and in CF sputum, and catabolized by P. aeruginosa. The beneficial effects of acetoin catabolism on both bacteria during their interaction highlighted trophic cooperation between P. aeruginosa and S. aureus in CF lung infections.
Materials and methods
Bacterial strains
The bacterial strains and plasmids used in this study are listed in Supplementary Tables S1 and S2. CF clinical strains were isolated by the Infectious Agents Institute from sputa of patients monitored in the two CF centres of Lyon (Hospices Civils de Lyon (HCL)). S. aureus and P. aeruginosa strains were isolated from patients co-infected by both bacteria. Each strain pair indicated in Supplementary Table S1 was recovered from a single sample, obtained from different patients in most cases, as indicated in Supplementary Table S1. All the methods were carried out in accordance with relevant French guidelines and regulations. This study was submitted to the Ethics Committee of the HCL and registered under CNIL No. 17-216. All the patients were informed of the study and consented to the use of their data.
As schematized in Supplementary Fig. S1A, interaction state was determined for each pair by growth inhibition tests on tryptic soy agar (TSA) and in liquid cultures [18]. As previously described, coexistence was characterized by: (i) the absence of inhibition halo on agar tests, and (ii) similar growth in monocultures and co-cultures for 8 h [18].
The knockout ΔacoR and Δaco mutants were generated in the P. aeruginosa PA2600 strain via allelic exchange owing to suicide plasmids pEXG2ΔacoR and pEXG2Δaco constructed as described previously [21, 22]. Gentamicin (Euromedex) was used at final concentrations of 50 µg/ml. Detailed protocols are given in Supplementary Data.
Cultures conditions
Strains were grown in monoculture or co-culture in brain–heart infusion (BHI), as described by Briaud et al. [18]. Briefly, S. aureus, P. aeruginosa, Burkholderia cenocepacia, Stenotrophomonas maltophilia and Bacillus subtilis overnight pre-cultures were diluted to an OD600 of 0.1 in BHI and grown for 2 h30 at 37 °C and 200 r.p.m. Cultures were then diluted to an OD600 of 2 in fresh medium and 10 ml was mixed with 10 ml BHI for monocultures. Co-cultures were performed by mixing 10 ml of P. aeruginosa suspension with 10 ml of S. aureus, B. cenocepacia, S. maltophilia or B. subtilis suspension. For supernatant exposure, 10 ml of S. aureus supernatant from a 4-h culture was filtered on a 0.22-µM filter and added to 10 ml of P. aeruginosa suspension. Cultures were grown for 8 h at 200 r.p.m. and at 37 °C for transcriptomic studies. Long-term survival assays were performed by extending the incubation time of S. aureus and P. aeruginosa monocultures and co-cultures up to 5 days. Plating at days 0, 3 and 5 was performed on mannitol salt agar (MSA, BBLTM Difco) and cetrimide (DifcoTM) for S. aureus or P. aeruginosa counts, respectively. For growth monitoring in the presence of acetoin, minimal medium M63 (76 mM (NH4)2SO4, 500 mM KH2PO4, 9 µM FeSO4⋅7H2O, 1 mM MgSO4⋅7H2O) was inoculated with P. aeruginosa to an OD600 of 0.1 and incubated for 25 h at 37 °C and 200 r.p.m. Every 2 h during 10 h and at t = 24 h, acetoin was added to obtain a final concentration of 1.5 mM in the culture. Plating on TSA was performed at t = 0, 1 h after each acetoin addition and at t = 24 h.
Genome sequencing and annotation
Genome sequencing and annotation of PA2596 and PA2600 strains were performed as previously described [18]. In order to compare CDS from PA2596 and PA2600, PAO1 strain (NC_002745.2) was used as a reference. Protein sequences were compared and grouped using a similarity threshold of 95% through Roary (V3.8.2) [23]. Gene names and numbers were gathered from PAO1 and used as ID tags for common genes. For non-common genes, CDS from PA2596 and PA2600 were tagged with a specific name (gene number and name recovered from the UniprotKB database). Functional classification was performed using the KEGG (Kyoto Encyclopaedia of Genes and Genomes) database and by a manual literature check of each gene function. The complete genome sequences for the PA2596 and PA2600 strains were deposited in GenBank under the accession number GCA_009650455.1 and GCA_009650545.1.
Transcriptomic analysis
Supplementary Fig. S1 schematizes the global methodology used for the transcriptomic analysis. RNA-sequencing (RNAseq) analysis was performed on four pairs: the patient-specific pairs SA2597/PA2596 (competitive) and SA2599/PA2600 (coexisting), and the crossed pairs SA2599/PA2596 (competitive) and SA2597/PA2600 (coexisting). RNA extraction, cDNA library preparation and sequencing and read treatment were conducted as previously described [18]. Gene expression was considered as dysregulated when: (i) the fold change between co-culture and monoculture was at least 4-fold with an adjusted P value < 0.05, (ii) dysregulation was observed in the two pairs of strains with the same interaction state and (iii) the dysregulation was specific to either coexistence or competition state. RNAseq data that support our findings are available in the SRAdatabase under the BioprojectID: PRJNA562449, PRJNA562453, PRJNA554085, PRJNA552786, PRJNA554237 and PRJNA554233.
Confirmation of gene expression was achieved by quantitative reverse transcription PCR (RT-qPCR) as previously described on 14 or 21 coexisting pairs including the SA2599/PA2600 couple used in the RNAseq experiment (Supplementary Fig. S1) [18]. Housekeeping genes gyrB and rpoD were used as an endogenous control. Supplementary Table S3 lists the primers used and the target genes.
Acetoin and glucose dosages
Acetoin was quantified using a modified Voges–Proskauer test [24] optimized in 96-well microplates. Thirty-five microlitres of creatine (0.5% m/v, Sigma), 50 µl of α-naphtol (5% m/v, Sigma) and 50 µl of KOH (40% m/v, Sigma) were added sequentially to 50 µl of culture supernatant. The mix was incubated for 15 min at room temperature and optical density at 560 nm was read using spectrophotometry (Tecan Infinite Pro2000, Tecan-Switzerland). Glucose dosages were performed with a glucose (Trinder, GOPOD) assay kit (LIBIOS) in microplates. A measure of 185 µl of dosage reagent was added to 5 µl of culture supernatant and incubated at 37 °C for 5 min. Optical density at 540 nm was measured. Acetoin and glucose standards (Sigma) were performed in water, BHI or M63 according to the experiment.
Statistical analysis
Statistical analyses were performed using the Prism GraphPad 8.0 software (San Diego, CA). Differences in gene expression fold change and bacterial survival were studied using one-way analysis of variance (ANOVA) with Dunnett’s or Tukey’s post-test comparisons, as specified in the corresponding figures. Median acetoin and glucose concentrations were compared using Mann–Whitney tests or Kruskall–Wallis tests with Dunn’s correction when appropriate. Differences were considered significant when P values were <0.05.
Results
The P. aeruginosa transcriptome is affected by the presence of S. aureus
We studied the genetic expression of P. aeruginosa in the absence or presence of S. aureus in two contexts: when P. aeruginosa and S. aureus were in competition or when they were in coexistence. We thus performed RNAseq analyses using a competitive strain pair (SA2597/PA2596) and a coexisting one (SA2599/PA2600). Each pair was recovered from a single sample of a co-infected CF patient (Supplementary Table S1). As the nature of interaction was determined solely by P. aeruginosa [18], the pairs were crossed to study gene expression in additional competitive (SA2599/PA2596) and coexisting (SA2597/PA2600) pairs. The transcriptomic effect was therefore evaluated during co-cultures of two competitive and two coexisting strain pairs (Supplementary Fig. S1). P. aeruginosa gene expression was considered dysregulated when dysregulation was common to both co-cultures in comparison to monoculture. Each dysregulated gene was then associated with a functional class by carrying out a KEGG analysis (Fig. 1a and Supplementary Tables S4 and S5).
a Number of under-expressed (grey bars) and overexpressed (black bars) genes of P. aeruginosa during co-culture with S. aureus for competitive (left) or coexisting (right) pairs. PA2596 competition and PA2600 coexistence strains were cultivated in the absence or presence of SA2597 or SA2599, as described in Supplementary Fig. S1. RNAs were extracted after 4 h of culture and the RNAseq analysis was performed as described in the “Material and methods” section. A gene was considered as differentially expressed when the fold change was >|2log2| with an adjusted P value <0.05. Functional classification was performed using the KEGG database and the literature. b Fold change of 26 P. aeruginosa gene expression induced by co-culture with S. aureus during coexistence interaction. Twenty-one coexisting S. aureus–P. aeruginosa pairs were isolated from separate CF sputa recovered from 20 different patients. Each P. aeruginosa strain was cultivated in the absence or presence of its co-isolated S. aureus strain. RNAs were extracted after 4 h of culture and gene expression was assayed by RT-qPCR. A gene was considered as differentially expressed when the fold change was >|2|, indicated by dotted lines. Black lines indicate the median. Genes were tested in 14 (regular character) or 21 (bold character) strains. The list of the strains used is shown in Supplementary Table S1. Genes annotated with (*) were not identified as dysregulated during the RNAseq experiment.
Sixty-eight P. aeruginosa genes were dysregulated in co-culture in the context of competition, with a majority of down-regulated genes (79.4%; Fig. 1a). Fifteen genes involved in nitrogen metabolism and 19 genes involved in iron metabolism were down-regulated, making these two functional classes the most affected in competition. Among these genes, the nir and nos systems involved in denitrification as well as genes implicated in iron uptake and transport (isc and fec genes) were down-regulated (Supplementary Table S4). Other functional classes were less affected, as only four genes linked to carbon and amino acid metabolism (bauA, ddaR, gntK and arcD) and three genes encoding membrane and virulence factors (rfaD, PA2412 and cdrA) were dysregulated in the presence of S. aureus.
More genes were affected during coexistence interaction, as the dysregulation of 105 genes was observed in P. aeruginosa (Fig. 1a). In spite of a trend of up-regulation (56.4% of genes), we could observe the down-regulation of 11 genes involved in nitrogen metabolism. Among these, eight were also down-regulated in the context of competition, especially genes from the nir system. We can thus presume that the down-regulation of nir genes is not specific to coexistence or competition interaction states. Conversely, the dysregulations of iron metabolism-related genes appeared to be specific to competition strains, as only one gene (fhp) of this functional class was down-regulated in coexistence. However, coexistence specifically affected numerous genes belonging to functional classes of carbon and amino acid metabolism (18 and 23 genes) and membrane and virulence factors (15 genes). Concerning the latter class, a trend towards lower expression was observed, probably due to the down-regulation of membrane-associated factors, such as the flp-tad system (flp, tad and rcpC genes encoding Flp pilus) and the PA1874-1876 operon (encoding an efflux pump) (Supplementary Table S5).
The classes most affected in coexistence with S. aureus were related to P. aeruginosa energetic metabolism (Supplementary Fig. S2). We observed a down-regulation of genes coding for major pentose phosphate pathway enzymes, like the gluconokinase GntK, its regulator GntR and the operon zwf-edaA (PA3183-PA3181), encoding a glucose-6-phosphate 1-dehydrogenase, a 6-phosphogluconolactonase and a 2-keto-3-deoxy-6-phosphogluconate aldolase [25]. We also noticed the down-regulation of five other genes clustered near this operon and involved in the same pathway (edd and gapA genes) and glucose transport (gltB, gltF and gltK) [25, 26].
In contrast, an up-regulation of numerous genes involved in the utilization of alternative carbon sources as butanoate and amino acids was observed. The aco system, comprising the operon PA4148-PA4153 and the gene encoding its transcriptional regulator acoR (PA4147), was up-regulated in P. aeruginosa coexisting with S. aureus. This system has been described in P. aeruginosa PAO1 to be responsible for 2,3-butanediol and acetoin catabolism [27]. According to KEGG analysis, the up-regulated genes acsA (PA0887), PA2555 (acs family) and hdhA (PA4022) are also involved in the butanoate pathway and energy production from 2,3-butanediol and acetoin, as their products catalyse the production of acetyl-coA from acetaldehyde and acetate (Supplementary Fig. S2). Finally, 23 genes implicated in amino acid metabolism were up-regulated in P. aeruginosa in the presence of S. aureus. Most of them were linked to the catabolism of several amino acids (Supplementary Fig. S2). In particular, we observed the liu operon (PA2015-PA2012), the mmsAB operon (PA3569-PA3570) and the hut gene system (PA5097-PA5100) involved in leucine, valine and histidine catabolism, respectively.
In order to confirm these transcriptomic effects, we co-cultivated a set of P. aeruginosa–S. aureus coexistence CF pairs and performed RT-qPCR to evaluate P. aeruginosa gene expression in the presence of S. aureus (Fig. 1b). Each pair was isolated from a single sputum. Twenty-six genes were tested, including 19 identified as dysregulated during RNAseq analysis and belonging to the most impacted categories, that is, carbon and amino acid metabolism, and membrane and virulence factors. Most of the genes were tested in a total of 14 P. aeruginosa–S. aureus pairs; the expression of four of these genes was assessed in seven additional pairs to confirm the dysregulations observed. The different P. aeruginosa strains presented very different transcriptomic patterns during co-cultivation with S. aureus, especially for membrane-associated and virulence factor genes. We noticed an overexpression of rcpC, tadA, tadG and flp from the flp-tad system from 52.4 to 35.7% of the P. aeruginosa strains. We also tested seven additional genes encoding virulence factors previously described as involved in P. aeruginosa interaction with S. aureus as las, rhl, pch and pvd genes [7]. Regarding the latter genes, no clear effect of S. aureus co-cultivation was observed.
However, clearer transcriptomic patterns were observed for genes linked to carbon and amino acid metabolism, the two gene classes most impacted during the RNAseq experiment (Fig. 1b). We confirmed the up-regulation of liuA gene in 78.6% (11/14) of P. aeruginosa strains co-cultivated with S. aureus. We also confirmed the down-regulation of glucose metabolism genes in a high proportion of strains, ranging from 92.9% (13/14) for zwf, gltF and edd genes to 71.4% (10/14) for gntK gene. Finally, the up-regulation of genes involved in butanoate metabolism was confirmed for acoR (57.2%, 12/21), PA4148 (66.7%, 14/21), acoB (57.1%, 8/14) and PA4153 (64.3%, 9/14), suggesting an impact of co-culture on the whole aco system in P. aeruginosa. In view of these results, we focused our study on four genes: liuA and zwf, respectively involved in leucine and glucose catabolism, PA4148, the first gene of the aco operon and acoR, both responsible for acetoin catabolism.
The P. aeruginosa aco system is induced by S. aureus acetoin
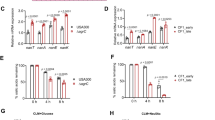
In order to determine if the transcriptomic dysregulations of acoR, PA4148, liuA and zwf in P. aeruginosa PA2600 are specific to interaction with S. aureus, we tested the effect of three other bacterial species: B. cenocepacia and S. maltophilia, as they are sometimes associated with P. aeruginosa in CF patients, and B. subtilis, as it produces a large amount of acetoin [28]. Dosages in B. cenocepacia and S. maltophilia monocultures confirmed that these bacteria do not produce acetoin (Supplementary Fig. S3) as previously described [29,30,31]. We observed a down-regulation of P. aeruginosa zwf gene expression in all co-cultures in comparison to monocultures, although the down-regulation induced by S. aureus was significantly greater (Padj < 0.02) (Fig. 2b). Regarding aco system genes (acoR and PA4148) and liuA gene, only B. subtilis induced levels of overexpression similar to S. aureus SA2599 (Fig. 2a, b). We thus hypothesized that acetoin, produced by these two species during our experiment (Supplementary Fig. S3), may be the inductor signal for these genes during co-culture with S. aureus.
a, b Fold changes of acoR and PA4148 (A) and liuA and zwf (b) induced by co-culture with S. aureus (black bars), B. subtilis (grey bars), B. cenocepacia (hatched white bars) or S. maltophilia (hatched black bars). P. aeruginosa PA2600 strain was cultivated in the absence or presence of S. aureus SA2599, B. subtilis, B. cenocepacia or S. maltophilia. RNAs were extracted after 4 h of culture and gene expression was assayed by RT-qPCR. Bars represent the mean fold change + SEM from three independent experiments. Dotted lines indicate a fold change = |2|. *Padj < 0.05, **Padj < 0.01 and ***Padj < 0.001 ANOVA with Dunnett’s correction (S. aureus vs. condition). c, d Fold changes of acoR and PA4148 (c) and liuA and zwf (d) induced by culture in supernatant of S. aureus UAMS-1 wild type (WT, black bars), ΔalsSD mutant (white bars) and ΔalsSD mutant complemented with increasing acetoin concentrations (grey bars). P. aeruginosa PA2600 strain was cultivated in the absence or presence of filtered supernatant from S. aureus UAMS-1 WT, ΔalsSD or ΔalsSD complemented with acetoin concentrations ranging from 0.375 to 2 mM. RNAs were extracted after 4 h of culture and gene expression was assayed by RT-qPCR. Bars represent the mean fold change + SEM from three independent experiments. The dotted line indicates a fold change = |2|. *Padj < 0.05 and **Padj < 0.01 ANOVA with Tukey’s correction.
In order to test this hypothesis, we first explored whether an inductor signal was present in the supernatant of S. aureus culture. We indeed observed an overexpression of acoR and PA4148 when P. aeruginosa PA2600 was cultivated in S. aureus SA2599 culture supernatant, as well as the down-expression of zwf, but to a lesser extent in comparison to co-culture (Supplementary Fig. S4). On the contrary, we did not observe overexpression of the liuA gene, suggesting that this effect is not due to acetoin and requires the presence of S. aureus cells. Second, we cultivated P. aeruginosa PA2600 in the presence of S. aureus UAMS-1 wild-type (WT) supernatant or its ΔalsSD derivative defective in acetoin synthesis [32]. The supernatant of the WT UAMS-1 strain induced the same transcriptomic patterns on P. aeruginosa as those obtained with the CF SA2599 strain (Supplementary Fig. S4). In the presence of UAMS-1 ΔalsSD supernatant, the overexpression of aco genes was almost totally eliminated (P = 0.0150 for PA4148) (Fig. 2c). However, the addition of acetoin to this supernatant restored this overexpression in a dose-dependent manner (P = 0.0020 for 0 vs. 1.5 mM for PA4148), but with threshold effects between 0.375 and 1.5 mM of acetoin (Fig. 2c). This indicates that induction through acetoin is one of the mechanisms involved in aco system overexpression in P. aeruginosa. This experiment showed that zwf gene down-regulation did not seem to be mediated by acetoin (Fig. 2d).
Coexisting isolates of S. aureus and P. aeruginosa efficiently metabolize acetoin
As the aco system is involved in acetoin catabolism in P. aeruginosa [27] and S. aureus produces this molecule [27, 28], we hypothesized that P. aeruginosa could catabolize S. aureus acetoin. To explore this hypothesis, we monitored acetoin concentration during monocultures and co-cultures of three pairs of strains: SA2599/PA2600 (Fig. 3a), SA146/PA146 and SA153/PA153A (Supplementary Fig. S5A, B). P. aeruginosa strains did not produce acetoin, but acetoin accumulation up to 1.3 mM was observed in S. aureus monocultures. Interestingly, a reduction of acetoin accumulation of at least 30% was observed when S. aureus was co-cultivated with P. aeruginosa, in comparison to S. aureus monoculture (Padj = 0.0172 for the pair SA2599/PA2600). The same trend could be observed in CF patient sputa, as a higher acetoin concentration was detected in sputa from S. aureus mono-infected patients (n = 9) in comparison to sputa from S. aureus–P. aeruginosa co-infected patients (n = 11), although it was not significant (Supplementary Fig. S6). This effect could be due to a down-regulation of S. aureus acetoin biosynthesis, or acetoin catabolism in co-culture. Growth of P. aeruginosa PA2600 in S. aureus SA2599 supernatant containing acetoin actually led to a reduction of acetoin concentration (P = 0.0247), demonstrating the ability of P. aeruginosa to catabolize acetoin (Fig. 3c). This ability was confirmed for the two other strains PA146 and PA153A when grown in the supernatant of SA146 and SA153, respectively (Supplementary Fig. S5C, D). We also noted that acetoin catabolism started after glucose depletion in the supernatant and could be delayed by glucose addition (Fig. 3c). This suggests that P. aeruginosa uses acetoin as an alternative carbon source in the absence of easily available substrates such as glucose. The early glucose depletion observed during co-culture supports this hypothesis (Fig. 3a).
a, b S. aureus SA2599 and P. aeruginosa PA2600 were cultivated in monoculture or co-culture. Acetoin (black) and glucose (grey) were quantified from the supernatant each hour. A Points represent the mean acetoin or glucose concentration ± SEM from three independent experiments. b Bars represent the mean area under the curves presented in a + SEM. *Padj < 0.05, **Padj < 0.01 and ****Padj < 0.0001 ANOVA with Tukey’s correction. Similar acetoin accumulation results are shown in Supplementary Fig. S5 for couples 146 and 153A. c, d A filtered supernatant of S. aureus SA2599 was inoculated with P. aeruginosa PA2600 culture or sterile medium for controls. The supernatant was used unaltered (dotted lines) or complemented with glucose (solid lines). Acetoin (black) and glucose (grey) were quantified from the supernatant each hour. c Points represent the mean acetoin or glucose concentration ± SEM from three independent experiments. d Bars represent the mean area under the curves presented in c + SEM. *P < 0.05 and ***P < 0.001 Student’s t test. Similar results of acetoin catabolism are shown in Supplementary Fig. S5 for couples 146 and 153A.
In order to test if this acetoin metabolism was specific to the interaction state between S. aureus and P. aeruginosa, we evaluated acetoin production and catabolism for 12 couples of competition and 12 couples of coexistence (Supplementary Table S1). Cultivated in P. aeruginosa supernatant, S. aureus strains from coexisting pairs were able to produce four times more acetoin (230 µM) than strains from competitive pairs (60 µM) (P < 0.0001) (Fig. 4a). This distinction was not observed during culture in rich medium (Supplementary Fig. S7). Regarding P. aeruginosa, we cultivated the sets of competitive and coexisting P. aeruginosa strains in SA2599 supernatant and monitored acetoin catabolism (Fig. 4b). Both competitive and coexisting strains catabolized acetoin since we observed a decrease in acetoin concentration for both groups (Padj = 0.0004 for coexistence). However, coexisting strains showed increased catabolism efficiency. Indeed, this group catabolized 98.6% of acetoin after 4 h of culture, while competitive strains catabolized only 47% of acetoin (Padj = 0.0315). This increased efficiency of acetoin production and catabolism for coexisting strains could not be explained by a difference in glucose utilization between competition and coexistence strains, as both groups catabolized glucose with the same efficiency (Supplementary Fig. S8). Acetoin production by S. aureus and catabolism by P. aeruginosa therefore seems to be more efficient in isolates from coexisting couples.
a Acetoin concentration in P. aeruginosa supernatant inoculated with S. aureus strains from competition and coexistence couples. Each S. aureus strain from competition (n = 12, grey bar) and coexistence (n = 12, black bar) couples was cultivated in P. aeruginosa PA2600 filtered supernatant and acetoin was quantified from the supernatant after 6 h of culture. Bars represent the median acetoin concentration ± 95% CI. ****P < 0.0001 Mann–Whitney test. b Acetoin concentration in S. aureus supernatant inoculated with P. aeruginosa strains from competition and coexistence couples. Each P. aeruginosa strain from competition (n = 12, grey bar) and coexistence (n = 12, black bar) couples was cultivated in S. aureus SA2599 filtered supernatant and acetoin was quantified from the supernatant after 4 h of culture. Sterile supernatant was used as control (white bar) for acetoin degradation. Bars represent the median acetoin concentration ± 95% CI. The dotted line indicates the initial acetoin concentration. *Padj < 0.05, ***Padj < 0.001 Kruskall–Wallis test with Dunn’s correction.
Acetoin catabolism by P. aeruginosa increases survival rates of both pathogens in co-culture
As P. aeruginosa catabolizes acetoin when medium is glucose-depleted, we tested the effect of acetoin on PA2600 growth in minimal medium M63 (containing no glucose or amino acids) supplemented with 1.5 mM acetoin every 2 h (Fig. 5). P. aeruginosa was able to grow up to 1.6 × 109 colony-forming unit (CFU)/ml after 24 h of culture with acetoin as sole carbon source, while PA2600 ΔacoR and Δaco mutants grew significantly less, reaching a maximum cell concentration of 1.5 × 108 CFU/ml at 24 h (Fig. 5a). In parallel, we quantified acetoin concentration. We observed an accumulation of acetoin throughout the experiment in the presence of PA2600 ΔacoR and Δaco mutants. For the WT strain, the accumulation of acetoin reached its maximum at 5 h of culture and afterwards slowly decreased to reach undetectable values at 10 h of culture, demonstrating the consumption of acetoin in such conditions (Fig. 5b). A delay of 3 h between the start of acetoin catabolism and the increased growth of the WT strain was noticed, possibly due to the adaptation of metabolism. Acetoin catabolism nonetheless promoted a 10-fold increase in growth of P. aeruginosa during extended culture in glucose-depleted medium.
P. aeruginosa PA2600 WT (black lines), ΔacoR (dark grey lines) and Δaco (light grey lines) strains were grown in M63 medium and 1.5 mM acetoin was added every 2 h, indicated by black arrows. a Cultures were plated on TSA each 2 h to count bacteria. Points represent the mean bacterial concentration ± SEM from three independent experiments. b Acetoin was quantified from the supernatant each hour. Points represent the mean acetoin concentration ± SEM from three independent experiments.
In order to assess the impact of acetoin catabolism on the survival of both pathogens, we co-cultivated P. aeruginosa PA2600, ΔacoR and Δaco mutants with S. aureus SA2599. As S. aureus was not able to grow in M63-poor medium, and since acetoin affects its long-term survival [33, 34], we performed a long-term culture (5 days) in BHI medium. We determined the survival rate of S. aureus co-cultivated with P. aeruginosa in comparison to monoculture (Fig. 6a). Co-culture with the WT strain of P. aeruginosa induced a S. aureus survival rate of 4.7 × 10−1 after 3 days of culture and of 5.1 × 10−3 after 5 days. S. aureus survival thus appears to be highly affected by long-term co-culture with P. aeruginosa, even if the strains coexist stably during shorter spans of culture [18]. The state of interaction with S. aureus thus seems to rely on nutritional conditions, a fact already observed elsewhere [35, 36]. However, the survival rate of S. aureus was even lower during co-culture with P. aeruginosa ΔacoR and Δaco mutants, reaching only 9.7 × 10−2 at 3 days of culture and 5.6 × 10−4 at 5 days. S. aureus survival was thus four to ten times lower when P. aeruginosa was not able to catabolize acetoin, in comparison to co-culture with a strain that catabolize the molecule efficiently. These results suggest that the accumulation of acetoin may impact S. aureus survival during co-culture with P. aeruginosa.
S. aureus SA2599 was cultivated in monoculture (white bars) or in the presence of P. aeruginosa PA2600 WT (black bars), ΔacoR (dark grey bars) and Δaco (light grey bars) during 5 days. Cultures were plated at days 0, 3 and 5 on MSA and cetrimide to count S. aureus and P. aeruginosa, respectively, and acetoin was quantified from supernatant. a, b Survival rate was estimated by dividing the bacterial concentration in co-culture by bacterial concentration in monoculture for each bacterium. Bars represent the mean survival rate + SEM from five independent experiments. *Padj < 0.05, ****Padj < 0.0001 one-way ANOVA with Dunnett correction (WT vs. condition). c Acetoin concentration was normalized to S. aureus counts. Bars represent the median acetoin concentration per 106 S. aureus ± 95% CI from five independent experiments. *Padj < 0.05, **Padj < 0.01, Kruskal–Wallis with Dunn’s correction.
In parallel, we determined the survival rate of P. aeruginosa (Fig. 6b). We noticed that in such conditions, the P. aeruginosa WT strain presented a growth advantage in co-culture, with a maximum 6.6-fold increase of the P. aeruginosa population after 5 days in the presence of S. aureus in comparison to monoculture (Fig. 6b). The opposite effect was observed for aco mutant strains, as their populations were reduced by 75% for ΔacoR and 32% for Δaco after 3 days of co-culture in comparison to monoculture, confirming the role of acetoin catabolism in favouring P. aeruginosa growth and survival.
In order to figure out if acetoin accumulation was involved in the decreased survival rate of S. aureus, we monitored acetoin concentration in S. aureus monoculture and co-cultures (Fig. 6c). As expected, we did not detect acetoin in co-culture with the PA2600 WT strain over the 5 days, but we observed an significant accumulation of acetoin with PA2600 ΔacoR and Δaco mutants (Padj < 0.002). The proportion of acetoin in co-culture (1300–1700 µM/106 S. aureus) was >500 times higher than in monoculture (2.5 µM/106 S. aureus) (Fig. 6c). We thus cultivated S. aureus in different acetoin/cells proportions and observed that acetoin had an inhibitory dose-dependent effect on S. aureus growth from 20 µM/106 S. aureus (Padj < 0.0001) (Supplementary Fig. S9). We concluded that acetoin accumulation may be responsible for decreased S. aureus survival during co-culture with PA2600 ΔacoR and Δaco mutants.
Taken together, these results demonstrated that acetoin catabolism improves S. aureus survival in co-culture, in comparison to co-culture with strains unable to catabolize acetoin, since high concentration of acetoin appears to impair S. aureus growth. In parallel, acetoin catabolism promotes P. aeruginosa survival as a nutritional alternative carbon source.
Discussion
Co-infection with S. aureus and P. aeruginosa is a frequent situation, especially in the lungs of CF patients, where co-infection accounts for 25–50% of cases [17,18,19]. In this context of co-infection, two states of interaction between the two pathogens have been described: the well-known competitive interaction where P. aeruginosa is able to inhibit the growth of S. aureus and the coexistence state where the growth of both species is not affected by either individual species. The first state was studied extensively and the leading bacterial determinants of S. aureus growth inhibition were described [7]. On the contrary, little is known about the impact of the state of coexistence on bacterial physiology. In the present study, we explored the impact of S. aureus on P. aeruginosa gene expression and physiology.
Comparing competitive and coexistence states, we observed that the down-regulation of the genes involved in iron metabolism was specific to competition. Most of these, such as fec genes and PA4467-PA4471 operon, are involved in ferrous iron uptake and down-regulated during iron-replete conditions [37,38,39]. These conditions are certainly generated by the lysis of S. aureus that provides an iron source to P. aeruginosa during competitive interaction [7, 36], a situation not observed in coexistence. This hypothesis is supported by the work of Tognon et al. [40] that also identified the down-regulation of iron metabolism genes during competitive interaction. Interestingly, they also noted typical responses of amino acid starvation, including the down-regulation of genes involved in branched-chain amino acid degradation in competitive P. aeruginosa. While we did not identify such dysregulation in competition, an overexpression of numerous genes involved in amino acid catabolism was noted during coexistence (Supplementary Table S5), emphasizing that these dysregulations depend on interaction.
More interestingly, we observed that both carbon and amino acid metabolism was specifically affected during coexisting interaction. Many genes involved in glucose catabolism were down-regulated in coexisting isolates during co-culture with S. aureus, especially when the medium was glucose-depleted (Fig. 3). It is noteworthy that the zwf gene, down-regulated in almost all the P. aeruginosa strains tested, encodes a glucose-6-phosphate dehydrogenase that converts glucose-6-phosphate to 6-phosphogluconate; the first enzyme in the Entner–Doudoroff pathway, which is central to carbon metabolism in Pseudomonas sp.
In this condition of glucose depletion, we demonstrated that P. aeruginosa was able to use an alternative carbon source provided by S. aureus: acetoin. Acetoin is a four-carbon molecule produced by the decarboxylation of α-acetolactate. Owing to its neutral nature, the production and excretion of acetoin during exponential growth prevents over acidification of the cytoplasm and the surrounding medium. When other carbon sources are exhausted, it can constitute an external energy source for fermentive bacteria [28].
Acetoin produced by S. aureus was shown to be an inductor of the aco operon and acoR expression in P. aeruginosa (Fig. 2c) [27], allowing acetoin catabolism. This occurred in the absence of glucose and was potentially mediated by carbon catabolic repression (Fig. 3), a situation that was already described in other bacteria such as B. subtilis [28]. However, threshold effects in acetoin-mediated induction and variability in aco system overexpression in P. aeruginosa strains (Figs. 1b and 2c) suggest that other regulatory mechanisms may be involved. Our study may also support the relationship between acetoin and branched-chain amino acid pathways. Indeed, the biosynthesis pathways of acetoin and leucine are co-regulated and share the same precursor α-acetolactate in S. aureus [28]. In response to co-culture with S. aureus, P. aeruginosa clinical strains showed overexpression of acetoin and leucine catabolism genes (Figs. 1b and 2), suggesting the presence of both compounds in our co-culture conditions. All of our analyses were performed in vitro. However, using Voges–Proskauer dosage, we were able to confirm the presence of acetoin in CF patient sputa, and in lower concentrations for P. aeruginosa-positive samples (Supplementary Fig. S6). No direct correlation between the presence of P. aeruginosa presence and the quantity of acetoin can be established as other microorganisms present in sputa may also have an impact on acetoin concentration. Nevertheless, our data support the work of Španěl et al. [41] and suggest that P. aeruginosa may catabolize and use S. aureus acetoin in the lung environment.
More importantly, we observed that the catabolism of acetoin by P. aeruginosa and acetoin production by S. aureus were both more efficient for coexisting isolates, in comparison to competitive ones (Fig. 4). This underlines the adapted metabolic regulation in coexisting isolates in comparison to competitive ones. It is well known that the coexistence phenotype between P. aeruginosa and S. aureus is a consequence of an adaptation process. Indeed, P. aeruginosa strains isolated from early infection outcompete S. aureus, while strains isolated from chronic infection are less antagonistic and can be co-cultivated with S. aureus [7, 10, 17]. It has also been widely described how both pathogens evolve during colonization to evade the immune response and antibiotic treatment [42]. Here, for the first time, we suggest that an evolutionary process leads to an adaptation of interspecies metabolic pathways between P. aeruginosa and S. aureus.
Therefore, we suggest that acetoin produced by S. aureus could contribute to sputum nutritional richness and be used by P. aeruginosa to survive in this nutritionally competitive environment during chronic infection. This hypothesis is supported by the beneficial effect of acetoin catabolism on P. aeruginosa growth and survival, especially during co-culture with S. aureus (Figs. 5 and 6b). S. aureus survival in the presence of P. aeruginosa was also shown to be highly affected by nutrient availability induced by co-culture conditions (Fig. 6a). While coexistence is characterized by an absence of S. aureus growth inhibition during 8-h co-culture [18], it appears that nutritional competition can still occur under unfavourable conditions and affect S. aureus’s survival. Therefore, coexistence between the two pathogens is promoted in nutritionally rich environments, in line with previous observations [35, 36]. Although its survival rate is affected under adverse nutritional conditions induced by long-term culture, acetoin catabolism benefits its producer, S. aureus (Fig. 6a). While this effect seems to be linked to acetoin accumulation in the medium, as demonstrated by P. aeruginosa ΔacoR and Δaco mutants that do not catabolize acetoin anymore (Figs. 5b and 6c), the precise mechanism remains unclear. In S. aureus, cell death in the stationary phase may be induced by acetate production and ensuing intracellular acidification. Thomas et al. [34] showed that acetoin production counters cytoplasmic acidification by consuming protons and promotes S. aureus’s survival in the late-stationary phase. We hypothesize that acetoin accumulation in the medium may induce a negative control of acetoin synthesis, affecting S. aureus’s survival during co-culture conditions.
Previous studies demonstrated the potential benefits of S. aureus and P. aeruginosa during co-infection. For example, S. aureus facilitates the survival of P. aeruginosa lasR mutants commonly found in CF patients by detoxifying surrounding nitric oxide released by host immune cells [43]. On the other hand, 4-hydroxy-2-heptylquinoline-N-oxide produced by P. aeruginosa cells inhibits respiration in S. aureus and also protects S. aureus cells from aminoglycosides [44]. In addition, we recently demonstrated that S. aureus antibiotic resistance and internalization into epithelial cells were increased in the presence of coexisting P. aeruginosa [18]. Here, we show that carbon metabolism is largely affected and that P. aeruginosa uses the acetoin produced by S. aureus as an alternative carbon source. This metabolic dialogue between the two pathogens is selected during bacterial adaptation in CF lungs and promotes their survival. Thus, we highlight for the first time trophic cooperation between S. aureus and P. aeruginosa during cooperative interaction.
References
Sibley CD, Rabin H, Surette MG. Cystic fibrosis: a polymicrobial infectious disease. Future Microbiol. 2006;1:53–61.
Guss AM, Roeselers G, Newton ILG, Young CR, Klepac-Ceraj V, Lory S, et al. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J. 2011;5:20–29.
Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev. 2012;25:193–213.
Murray JL, Connell JL, Stacy A, Turner KH, Whiteley M. Mechanisms of synergy in polymicrobial infections. J Microbiol Seoul Korea. 2014;52:188–99.
Abisado RG, Benomar S, Klaus JR, Dandekar AA, Chandler JR. Bacterial quorum sensing and microbial community interactions. mBio. 2018;9:e02331–17.
Tashiro Y, Yawata Y, Toyofuku M, Uchiyama H, Nomura N. Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. 2013;28:13–24.
Hotterbeekx A, Kumar-Singh S, Goossens H, Malhotra-Kumar S. In vivo and in vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front Cell Infect Microbiol. 2017;7:106.
Serra R, Grande R, Butrico L, Rossi A, Settimio UF, Caroleo B, et al. Chronic wound infections: the role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev Anti Infect Ther. 2015;13:605–13.
O’Brien TJ, Welch M. Recapitulation of polymicrobial communities associated with cystic fibrosis airway infections: a perspective. Future Microbiol. 2019;14:1437–50.
Baldan R, Cigana C, Testa F, Bianconi I, De Simone M, Pellin D, et al. Adaptation of Pseudomonas aeruginosa in cystic Fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS ONE. 2014;9:e89614.
Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–92.
Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, Ramsey BW, et al. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros J Eur Cyst Fibros Soc. 2009;8:66–70.
Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol. 2012;10:841–51.
Marvig RL, Sommer LM, Molin S, Johansen HK. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet. 2015;47:57–64.
Limoli DH, Whitfield GB, Kitao T, Ivey ML, Davis MR, Grahl N, et al. Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. mBio. 2017;8:e00186–17.
La Rosa R, Johansen HK, Molin S. Adapting to the airways: metabolic requirements of Pseudomonas aeruginosa during the infection of cystic fibrosis patients. Metabolites. 2019;9:234.
Michelsen CF, Christensen A-MJ, Bojer MS, Høiby N, Ingmer H, Jelsbak L. Staphylococcus aureus alters growth activity, autolysis, and antibiotic tolerance in a human host-adapted Pseudomonas aeruginosa lineage. J Bacteriol. 2014;196:3903–11.
Briaud P, Camus L, Bastien S, Doléans-Jordheim A, Vandenesch F, Moreau K. Coexistence with Pseudomonas aeruginosa alters Staphylococcus aureus transcriptome, antibiotic resistance and internalization into epithelial cells. Sci Rep. 2019;9:16564.
Briaud P, Bastien S, Camus L, Boyadijian M, Reix P, Mainguy C, et al. Impact of coexistence phenotype between Staphylococcus aureus and Pseudomonas aeruginosa isolates on clinical outcomes among cystic fibrosis patients. Front Cell Infect Microbiol. 2020;10:266.
Frydenlund Michelsen C, Hossein Khademi SM, Krogh Johansen H, Ingmer H, Dorrestein PC, Jelsbak L. Evolution of metabolic divergence in Pseudomonas aeruginosa during long-term infection facilitates a proto-cooperative interspecies interaction. ISME J. 2016;10:1323–36.
Carriel D, Simon Garcia P, Castelli F, Lamourette P, Fenaille F, Brochier-Armanet C, et al. A novel subfamily of bacterial AAT-fold basic amino acid decarboxylases and functional characterization of its first representative: Pseudomonas aeruginosa LdcA. Genome Biol Evol. 2018;10:3058–75.
Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2005;102:8006–11.
Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinforma Oxf Engl. 2015;31:3691–3.
Nicholson WL. The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Appl Environ Microbiol. 2008;74:6832–8.
Lessie TG, Phibbs PV. Alternative pathways of carbohydrate utilization in pseudomonads. Annu Rev Microbiol. 1984;38:359–88.
Chevalier S, Bouffartigues E, Bodilis J, Maillot O, Lesouhaitier O, Feuilloley MGJ, et al. Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol Rev. 2017;41:698–722.
Liu Q, Liu Y, Kang Z, Xiao D, Gao C, Xu P, et al. 2,3-Butanediol catabolism in Pseudomonas aeruginosa PAO1: 2,3-butanediol catabolism in Pseudomonas aeruginosa. Environ Microbiol. 2018;20:3927–40.
Xiao Z, Xu P. Acetoin metabolism in bacteria. Crit Rev Microbiol. 2007;33:127–40.
Gade N, Negi SS, Jindal A, Gaikwad U, Das P, Bhargava A. Dual lower respiratory tract infection by Burkholderia cepacia and Acinetobacter baumannii in a neonate: a case report. J Clin Diagn Res. 2016;10:DD01–DD03.
Amoli RI, Nowroozi J, Sabokbar A, Rajabniya R. Isolation of Stenotrophomonas maltophilia from clinical samples: an investigation of patterns motility and production of melanin pigment. Asian Pac J Trop Biomed. 2017;7:826–30.
Dryahina K, Sovová K, Nemec A, Španěl P. Differentiation of pulmonary bacterial pathogens in cystic fibrosis by volatile metabolites emitted by their in vitro cultures: Pseudomonas aeruginosa, Staphylococcus aureus, Stenotrophomonas maltophilia and the Burkholderia cepacia complex. J Breath Res. 2016;10:037102.
Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS ONE. 2008;3:e3361.
Chaudhari SS, Thomas VC, Sadykov MR, Bose JL, Ahn DJ, Zimmerman MC, et al. The LysR-type transcriptional regulator, CidR, regulates stationary phase cell death in Staphylococcus aureus: metabolic control of cell death in S. aureus. Mol Microbiol. 2016;101:942–53.
Thomas VC, Sadykov MR, Chaudhari SS, Jones J, Endres JL, Widhelm TJ, et al. A central role for carbon-overflow pathways in the modulation of bacterial cell death. PLoS Pathog. 2014;10:e1004205.
Miller CL, Van Laar TA, Chen T, Karna SLR, Chen P, You T, et al. Global transcriptome responses including small RNAs during mixed-species interactions with methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa. MicrobiologyOpen. 2017;6:e427.
Mashburn LM, Jett AM, Akins DR, Whiteley M. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol. 2005;187:554–66.
Visca P, Imperi F. An essential transcriptional regulator: the case of Pseudomonas aeruginosa Fur. Future Microbiol. 2018;13:853–6.
Cornelis P, Matthijs S, Van Oeffelen L. Iron uptake regulation in Pseudomonas aeruginosa. Biometals Int J Role Met Ions Biol Biochem Med. 2009;22:15–22.
Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol Microbiol. 2002;45:1277–87.
Tognon M, Köhler T, Luscher A, van Delden C. Transcriptional profiling of Pseudomonas aeruginosa and Staphylococcus aureus during in vitro co-culture. BMC Genomics. 2019;20:30.
Španěl P, Sovová K, Dryahina K, Doušová T, Dřevínek P, Smith D. Do linear logistic model analyses of volatile biomarkers in exhaled breath of cystic fibrosis patients reliably indicate Pseudomonas aeruginosa infection? J Breath Res. 2016;10:036013.
Baishya J, Wakeman CA. Selective pressures during chronic infection drive microbial competition and cooperation. NPJ Biofilms Microbiomes. 2019;5:16.
Hoffman LR, Richardson AR, Houston LS, Kulasekara HD, Martens-Habbena W, Klausen M, et al. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog. 2010;6:e1000712.
Hoffman LR, Déziel E, D’Argenio DA, Lépine F, Emerson J, McNamara S, et al. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2006;103:19890–5.
Acknowledgements
This work was supported by the Fondation pour la Recherche Médicale, grant number ECO20170637499 to LC; the Finovi foundation to KM; the associations “Vaincre la mucoviscidose” and “Gregory Lemarchal” to KM. We thank Kenneth W Bayles from the University of Nebraska Medical Center (Omaha) for providing S. aureus UAMS-1 WT and mutant strains.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
All the strains used in this study were collected as part of the periodic monitoring of patients at the Hospices Civils de Lyon (HCL). This study was submitted to the Ethics Committee of the HCL and registered under CNIL No. 17-216. All the patients were informed of the study; however, as the study was non-interventional, no written informed consent was required under local regulations.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Camus, L., Briaud, P., Bastien, S. et al. Trophic cooperation promotes bacterial survival of Staphylococcus aureus and Pseudomonas aeruginosa. ISME J 14, 3093–3105 (2020). https://doi.org/10.1038/s41396-020-00741-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-00741-9