Abstract
Lytic viruses are believed to affect both flow patterns and host diversity in microbial food webs. Models resolving host and virus communities into subgroups can represent both aspects. However, when flow pattern is the prime interest, such models may seem unnecessary complex. This has led to proposals of black-box models using only total community sizes as state variables. This simplification creates a coexistence problem, however, since predator and virus communities then compete for the same, shared, prey = host community. Mathematically, this problem can be solved by introducing feedbacks allowing community-level properties to adapt. The different mathematical alternatives for such feedback represent different ecological assumptions and thus different hypotheses for how the balance between predators and viruses is controlled in nature. We here explore a model where the feedback works through an increase in host community resistance in response to high virus abundances, thereby reducing virus production. We use a dynamic “strategy” index S to describe the balance between defensive and competitive abilities in the host community, and assume the rate of change in S to be proportional to the local slope of the per capita fitness gradient for the host. We explore how such a “grey-box” model can allow stable coexistence of viruses and predators, and how equilibrium food web structure, virus-to-host ratio, and partitioning of host production varies; both as functions of host community traits, and as functions of external bottom-up and top-down drivers.
Similar content being viewed by others
Introduction
In contexts where the flow pattern for energy and elements through the pelagic food web is an issue, lytic viruses are believed to have an important role in shunting parts of the flow out of the predatory food chain. The result is a release of matter to the detrital and dissolved pools, from where large parts of the organic-C, nitrogen, and phosphorous are reincorporated into heterotrophic prokaryotes and/or phytoplankton [1, 2]. The quantitative role of these processes is believed to be significant, as e.g. estimates have suggested that bacterial production can be partitioned evenly between predation and viral lysis [3]. Models with an ambition to correctly represent the relationship between regenerated and new production in the photic zone therefore seem to need a dynamic representation of this partitioning.
In several models resolving the host community into host groups, viruses act as the mechanism that compensate for differences in growth rate between these groups [4,5,6,7]. Virus abundance, activity, and shunting of material out of the predatory pathway are then all driven by these growth rate differences in the host population; intimately linking ecosystem function to diversity. Models with no internal resolution of host diversity can obviously not mimic this mechanism in any direct manner. In recent versions, constructed to account for the combined role of competitive and defensive traits in host species diversity, host community is resolved to strain level [6]. Such high resolution may seem necessary to capture the link between biogeochemistry and diversity aspects of host–virus interactions, and also to incorporate the dynamics of evolutionary host–virus arms races. The large number of state variables, however, would make their direct application as dynamic modules in e.g. biogeochemical circulation models cumbersome and computationally costly. A tempting alternative is therefore to represent only the total size of host and virus communities with dynamic state variables. We will refer to such representations as black-box models. A recent example of this type of model can be found in Weitz et al. [8].
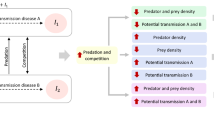
Food webs, as opposed to food chains, have points where the flows split due to competition for a shared resource. This creates a coexistence problem formulated as the general principle of competitive exclusion [9]. A black-box model where viruses (V) and predators (P) both attack a shared host (=prey) community (H) (Fig. 1a) is a special case of this. To illustrate the problem, one can calculate the host communities needed for production of viruses \(\left( {H_V^ \ast } \right)\) (Eq. 1, Box 1) and predators \(\left( {H_P^ \ast } \right)\) (Eq. 2) to equal their respective losses. The model in Fig. 1a has three types of parameters, the yields (Y-parameters) that express the fraction of limiting element transferred from prey/host to predator/virus, the clearance rate/adsorbtion coefficient (α-parameters) and the loss rates (δ-parameters) representing specific loss rates for predators and viruses. For the model in Fig. 1a to allow stable coexistence, a necessary condition is that the two top-down controls give the same result: \(H_V^ \ast = H_P^ \ast\), leading to:
Idealized Host–Virus–Predator food web used to illustrate the difference between a black-box models where stable coexistence of the three communities requires variability in one or more of the six Y, α or δ parameters, and b community-resolved models where viruses control host groups and steady state requires that the predator-controlled host community equals the sum of virus-controlled host groups
With all six parameter values fixed, this equality will not be valid in the general case. One will therefore need additional assumptions that introduce feedback mechanisms, making one or more of the Y, α, or δ parameters adaptive.
From Eq. 3, there are three broad classes of theoretically possible feedback mechanisms:
- (1)
Y-mechanisms, affecting the ratio between virus and predator yields.
- (2)
α-mechanisms, affecting the ratio between adsorption constant and clearance rate.
- (3)
δ-mechanisms, affecting the ratio between virus and predator loss rates.
The three classes represent very different physiological and ecological mechanisms: First, yields (Y) are the efficiency by which the limiting element (phosphorous in our model) is transferred from prey (host) to predator or virus. Changes in burst size would fall in this category. Second, maximum values for the adsorption constant and clearance rate (α) represent the encounter kernel dominated by diffusion for the host–virus interaction, but swimming for the predator–prey interaction [10], both potentially reduced by success rates <1 in such encounters, e.g. caused by defence mechanisms in the host population. The model explored here belongs to this class. Third, loss rates (δ) are probably dominated by inactivation processes for viruses, but by secondary predators for the predator. For each class, introduction of feedback into these parameters requires very different assumptions concerning physiological and/or ecological mechanisms, each providing a different potential solution to Hutchinson’s paradox. Theoretical mechanisms potentially contributing to the so far unexplained observation of high diversity in aquatic microbial communities may thus be sought in a diverse set of feedback mechanisms, acting in concert with other factors such as diversity in limiting substrates and inhomogeneities in space and time.
The resolved models have a different solution to this coexistence problem. Resolving the host community into host groups (Fig. 1b) HVi, I = 1..n. [5] allows the expected relationships \(H_{Vi}^ \ast \ll H_P^ \ast\) to be retained. The constraint in Eq. 3 is then replaced with the condition that total size of the predator-controlled host community must equal the sum of host group sizes (Eq. 4). Through this, richness (n) in host groups becomes an emergent property of the model. The black-box models’ need for parameter variability is thus replaced by the variable n. Increasing total host community size will here allow competitively inferior host groups (i.e. with lower growth rates µi) to establish. For illustration, Fig. 1b is arranged with decreasing µi from left to right. Increasing the community size will thus allow addition of new, competitively inferior, host groups with lower growth rates, but also lower loss rates, to establish at the right end.
The maximum adsorption constant of viruses (αV) is much higher than the clearance rate of predators (αP) [11]. Using this directly at community-level would lead to \(H_V^ \ast \ll H_P^ \ast\). With viruses able to reproduce on a much smaller host population than that required by predators, viruses would outcompete predators over time. A model where a single virus-community control an aggregated set of host groups like, e.g., the community of heterotrophic prokaryotes, would therefore need to be parameterized with some kind of community adsorption coefficient αV smaller than the theoretical maximum [12]. The relationship between the community-level adsorption constant αV used in Eq. 3, and group-level adsorbtion constants αVi in the resolved model, can be derived by inserting the total, virus-controlled community size \(H_V^ \ast = \frac{{\delta _V}}{{Y_V\alpha _V}}\) as the right side of Eq. 4 (Box 1):
To illustrate the difference between community and group-level adsorbtion constants, assume a negligible immune population \(H_n^ \ast \ll H_V^ \ast\), all δVi = δV, and all YVi = YV. We then have \(\frac{1}{{\alpha _V}} = \mathop {\sum}\nolimits_{i = 1}^{n - 1} {\frac{1}{{\alpha _{Vi}}}}\); or, if also all αiV are identical, this simply gives \(\alpha _V = \frac{{\alpha _{Vi}}}{{n - 1}}\). While sufficiently resolved (high n) models can use theoretical αVi – values derived from collision theory, community-level αV in black-box models should be smaller than this. Also, a variation of n in resolved models will correspond to a variable αV, even if the groups added or removed should have identical αVi. Coexistence in resolved models can thus be seen as a special case where Eq. 3 is satisfied through variability in αV.
Previous virus–predator black-box models [8] use a δ-mechanism in the predator food chain to solve the coexistence problem (see SI for further discussion). As an alternative that more closely resembles the within-community adaptations of the resolved models, we here suggest a model where coexistence is obtained using adaptive α-parameters.
An α-mechanism model
Formulation of an adaptive host strategy
In resolved models, the negative density control of group-specific viruses lead to structural changes in host community composition, generating the negative feedback where e.g. a high viral abundance favors more defensive species and strains, thereby reducing virus production. The feedback mechanism explored here mimics this type of feedback by reducing the host community’s vulnerability to viral infection in response to high viral abundances. To do this, we introduce a new state variable S that represents the balance between competitive and defensive abilities. For convenience, we will refer to S as “community strategy”, but there is of course no suggestion in this of any kind of community intent. The assumption is that, as individuals with higher fitness (net reproduction rate) become dominating within the community, this can be represented as an increase in per capita fitness for the community. Details of the underlying selection and mutation mechanisms are not specified in our model.
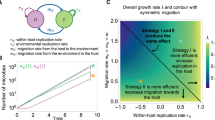
Following the formulations used by Våge et al. [13], community strategy S is defined as a dimensionless index 0 ≤ S ≤ 1. The value of S determines host nutrient affinity αH and virus adsorption αV through the relationships:
and
where τ is a parameter expressing trade-off between competition and defence (Fig. 2b).
Food web model with adaptive host strategies. a Structure of the nutrient—host(prey)–virus–predator food web model used. The food web is closed with respect to the limiting nutrient, and the total nutrient content NT and the top predator Z are used to study external bottom-up and top-down forcing, respectively. b Variation in the host community’s relative competitiveness c \(\left( { = \frac{{\alpha _H}}{{\alpha _H^0}}} \right)\) and vulnerability ν \(\left( { = \frac{{\alpha _V}}{{\alpha _V^0}}} \right)\), with host strategy S and trade-off τ
This gives a description where low S represents a community dominated by vulnerable competition strategists, shifting toward dominance of defence strategists unable to grow as S approaches 1 (For S = 0, relative competitiveness \(c = \frac{{\alpha _H}}{{\alpha _H^0}}\) and relative vulnerability \(v = \frac{{\alpha _V}}{{\alpha _V^0}}\) are both 1; for S = 1, they are both zero). For a trade-off τ < 1, large decreases in vulnerability is possible with little loss in competitiveness (Fig. 2b). If τ < 1, a small decrease in vulnerability will give a large loss in competitiveness. Note also that, for τ < 1, there is a strategy (S = 0.5) that maximizes the difference c − v.
Defining host community fitness as the net community growth rate, we assume that changes in S are in the direction of increasing fitness. Some of the properties of this model can be understood by looking at its steady state solution.
Approximate steady state solution
To illustrate the properties of the steady state, we will use the simplifying assumption of Holling Type I functional relationships (The simulations are run with Type II functions for the host and the predator).
Insertion of αV(S) and using \(\delta _P = \alpha _ZZ\) allows solving Eq. 3 for S to give the strategy Sco required for coexistence of viruses and predators as a function of external predator abundance Z:
With a Holling Type II response in the predator, the expression for Sco is slightly modified (Eq. 17, Box 2; see SI for details):
For stable coexistence, the equilibrium strategy S* must coincide with Sco.
In models with adaptive community-level traits, the slope of the relationship between an individual’s trait value and its fitness has been used as an approximation of the rate of change of a community-level trait [14]. Using net specific growth rate as the expression for host fitness (FH), we have:
with 1st and 2nd partial derivatives in the point N,V with respect to host strategy S:
and
respectively.
As a first approximation, we assume that the rate of change in S in the point N, V is proportional to the slope of the fitness gradient in this point:
The essential feedback mechanism in this model is that high viral numbers give an increase in S and the corresponding increase in defence leads to reduced virus production. The steady state can be calculated as follows (superscript“*” used to indicate steady state values).
For trade-offs τ < 1, the 2nd derivative is always <0, implying that for a given set of the two state variables N and V, there is not more than one extreme point where \(\frac{{\partial F}}{{\partial S}} = 0\) in the feasible range 0 < S < 1. If such an extreme point exists, it will be a maximum point, and the corresponding equilibrium strategy S* will also be the optimum strategy where\(\left( {\frac{{\partial F_H}}{{\partial S}}} \right)_{N^ \ast ,V^ \ast } = 0\). Equation 20 then gives:
Since S* must coincide with the strategy Sco that allows coexistence, rearranging Eq. 22 gives a constraint on the ratio between N* and V*:
At steady state, also net host growth rate (host fitness) must be zero:
Restricting this analysis to the case of a system with a closed nutrient budget we have
where NT is the total content of limiting element in the system and all pool sizes are in the same unit as this.
Inserting Eqs. 23, 24 and 2 (Box 1) in Eq. 25 and solving for V* gives:
Equation 26 illustrates how (in this food web), steady state viral abundance is a function both of the bottom-up driver NT and the top-down driver Z. Note that Z influences both the numerator and the denominator since Sco is a function of Z.
If locally stable, this equilibrium strategy represents the evolutionary stable community [15] for the food web context and environmental drivers given.
Replacing the Holling Type I with Type II functional relationships for host and predator, we have explored the dynamics of this model (Fig. 2a, Box 2) numerically. Simulations used to generate Figs. 4 and 5 have been run for 4000 h using the MatLab® ODE23 routine. The Matlab® script used to run the simulations and create Figs. 3–5 is included in the SI.
Standard Run (SR). Simulation of the Model in Fig. 2 with parameter values and initial conditions as given in Table 1. a Simulation of the transient shift in food web structure as a virus-free system near steady state is infected by viruses. b Changes in host community strategy S (solid line), here converging to the coexistence strategy Sco (broken line) allowing stable virus–predator coexistence
Changes in steady state structure of the food web in Fig. 2a as functions of the (upper row) community properties a trade-off τ and b maximum adsorption constant \({\mathrm{\alpha }}_V^0\), and (lower row) the external drivers c Z and d NT . Values used in the Standard Run (Fig. 3) are marked with dotted vertical lines. Only one parameter varied in each case, the other kept as in the Standard Run shown in Fig. 3. The y-axis in dimensionless units for S, in nM-P for the other state variables
Effects of (upper row) community properties a τ and b \({\mathrm{\alpha }}_V^0\), and (lower row) external drivers c Z and d NT on the relative importance of viruses versus hosts (VHR) and lysis versus predation (LPR) at steady state. Same simulation runs as in Fig. 4. Note logarithmic y-scale
Results
To explore the properties of this model, we have chosen one set of parameters (Table 1) and a standardized initial condition to produce a reference simulation run (Standard Run SR, Fig. 3). The standardized initial condition is obtained by calculating the (approximate, see SI for details) steady state without viruses and setting V and S to small values (Table 1). This to simulate viruses invading a virus-free system and to explore whether a new steady state with virus–predator coexistence is established. With the parameter set used, the transient response of SR is a damped oscillation toward steady state with most transients disappearing within ca. 4 weeks. Effects on the steady state were studied by varying a single parameter or external driver at a time. Equilibrium values for the state variables are shown as functions of two parameters characterizing the host–virus interaction (τ and \(\alpha _V^0\), Fig. 4a, b), and for the two external drivers Z and NT (Fig. 4c, d). The relative role of viruses at steady state in these simulations are further illustrated using the virus-to-host ratio (VHR*) and the lysis-to-predation ratio (LPR*) (Fig. 5a–d) for the same runs. To reveal cases not reaching steady state within 4000 h, both minimum and maximum value for the last 100 time points of each simulation are plotted in Figs. 4 and 5. As maximum and minimum values were identical for all simulations shown, this implies that for all parameter combinations used to generate Figs. 4 and 5, the viral invasion lead to a stable steady state solution. The steady state seemed to become locally unstable with persistent oscillations for values of Z and NT lower and higher, respectively, than the range used for these in Fig. 4, but the mechanisms behind these destabilizations have not been explored further. The oscillatory pattern is also influenced by the adaptation constant r, where a higher r lead to more rapid dampening of the transients (results not shown).
In these simulations, high absolute values for V* corresponded to high trade-off τ, as expected when defence is expensive (Fig. 4a). As also expected, V* increases with increasing maximum community adsorption constant \(\alpha _V^0\), but the effect saturates around the value used in SR. This is interesting as it means that, with the feedback in this model, more aggressive viruses with higher \(\alpha _V^0\) have little effect on ecosystem structure and function at steady state, but changes the internal composition of the host community toward more resistant phenotypes. Due to the top-down control from Z on host community size, H* increases linearly with Z (Fig. 4c) leaving less of NT for the three other variables. N*, and P* both decrease with increasing Z, while V* has a maximum point around the value used in SR, decreasing at both high and low Z. For fixed Z, H* is constant, and an increase in NT (Fig. 4d) leads to monotonic increase in all three of N*, P*, and V*. With this, equilibrium VHR* (Fig. 5) increases with increasing values of both τ, \(\alpha _V^0\), and NT, but decreases at high values of Z. The LPR* increases with τ, \(\alpha _V^0\), and Z (Fig. 5a–c), but decreases with NT (Fig. 5d).
Discussion
Black-box models where two communities, each represented as a single plankton functional type (PFT), compete for a single shared resource, will have two top-down controls for the shared resource. We have here explored how adaptation in the host’s defensive ability, subject to a cost of resistance in the form of decreased competitive ability, can function as such a feedback and allow stable coexistence between viruses and predators sharing the host (= prey) community as their single common limiting resource.
An additional feature of this model is the dynamic optimization of host fitness. Fitness optimization is conceptually attractive and adds only one extra parameter to the model (i.e. rate constant for community adaption, r), but is not the only formulation for the dynamics of S that can give stable steady states (results not shown).
In the more resolved models (Fig. 1b), coexistence emerges from flexibility in host community richness, a property not possible to represent in black-box models with fixed community traits. In our “grey-box” alternative, all internal shifts in physiology, any evolutionary arms races, and all internal population dynamic shifts in species and strains, is represented only by variation in the single strategy variable S. With the present lack of knowledge of these complex internal processes, the moderate number of assumptions needed in the grey-box representation is likely a great advantage in many applications.
With the feedback located in host defence, we get a model that can be top-driven. It is therefore possible to use this approach in contexts where the cascading effect from seasonal copepod migration has been hypothesized to be a central explanatory factor behind observed contrasts in bacterial and virus abundance, community composition, and population dynamics [16]. Such cascading effects seem difficult to reproduce with models where the feedback allowing coexistence is based on positive correlation between a predator biomass and its loss rate.
Microbes can defend themselves also against predation using mechanisms such as morphological changes [17,18,19] and toxic metabolites [20]. Importantly, some of these defence mechanism require organic-C. Predator defence in bacteria therefore becomes a function of whether bacterial growth is limited by organic-C or mineral nutrients [18, 21, 22]. An interesting extension of the approach explored here would therefore be a two-dimensional strategy, allowing for combined defence against viruses and predators. The associated 2D optimization problem would involve how system state controls bacterial limitation. Considering the rapid increase in plankton functional types when using resolved models to combine predator and viral defence [23], such a grey-box approach is likely an attractive alternative.
The central mechanism that allows coexistence in the model explored here is the effect of host adaptation on the effective adsorption constant αV and its trade-off based coupling to host affinity αH. The effective adsorption constant is a description of the host–virus interaction and therefore affected by evolutionary and population dynamic processes, not only in the host community, but also in viruses. The virus-side of host-virus arms races is not explicitly represented in our model. A model for host–virus arms races with adaptive dynamics has been developed by Weitz et al. [24] and a model of host evolution allowing more explicit formulation of the mechanisms involved has been studied by Menge and Weitz [25].
We have discussed the constraint represented by Eq. 3 as if the six parameters can vary independently. There are potential trade-offs between traits in both hosts [26, 27] and viruses [28, 29] and presumably also in the predators. The resulting potential for covariation between the six parameters in Eq. 3 has not been explored here.
Extensive virus - microbe data is now available [30, 31]. The framework outlined here seems to have a potential as a tool for deeper theoretical analysis of such data, but this requires integration of the simple food web used here into a more realistic representation of the microbial food web.
Ecological and evolutionary processes can be captured using adaptive dynamics in trait-based models [32]. We illustrated here that adaptive community strategies can be used as an alternative to multi-PFT models. This may have an advantage in terms of computational costs. More important for many applications is probably the potential it offers for retaining most of the transparency of relatively simple and well-understood few-PFT models.
The simple food web in Fig. 1a with a closed nutrient budget was chosen to illustrate the principles of coexistence due to host community adaptation with as few assumptions as possible. Using this approach in more complicated food web models will bring in additional coexistence issues such as phytoplankton–phytoplankton and phytoplankton–bacteria coexistence on a single limiting nutrient. These are explainable with the feedback from negative density control embedded in the “Killing-the-Winner” (KtW) mechanism [33]. Coexistence mechanisms seemingly belonging to different classes will then be combined in the same model. Intriguingly, however, the resolved models use the KtW mechanism to explain all these coexistences [33], suggesting that, at a deeper level they are rooted in conceptually similar negative density controls.
Models with a closed nutrient budget (constant NT) are important conceptually, and practically useful for enclosed systems like, e.g., mesocosms. If, however, such models are implemented as descriptions of the microbial food web in the pelagic photic zone, NT becomes a variable subject to nutrient import and export. A central external driver for these processes is then the mixing coefficient (fraction of photic zone water volume exchanged per unit time). The steady state condition becomes different as the classical import = export argument [34] can be reformulated to: The steady state nutrient content \(\left( {N_T^ \ast } \right)\) is the one that corresponds to a food web where export = import. Adding viruses to these food webs will affect the regenerated production and thus the f-ratio. An illustration of how mixing coefficient affects equilibrium food web structure in an open version of this model is included in SI, Figs. S1 and S2. To make this a realistic representation of pelagic viruses, however, a more complicated food web model including heterotrophic prokaryotes, their viruses, their phytoplankton competitors, micro- and mezo-zooplankton would be needed.
References
Riemann L, Middelboe M. Viral lysis of marine bacterioplankton: Implications for organic matter cycling and bacterial clonal composition. Ophelia. 2002;56:57–68.
Bratbak G, Jacobsen A, Heldal M. Viral lysis of Phaeocystis pouchetii and bacterial secondary production. Aquat Micro Ecol. 1998;16:11–6.
Fuhrman JA, Noble RT. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol Oceanogr. 1995;40:1236–42.
Thingstad TF, Lignell R. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat Micro Ecol. 1997;13:19–27.
Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr. 2000;45:1320–8.
Thingstad TF, Vage S, Storesund JE, Sandaa R-A, Giske J. A theoretical analysis of how strain-specific viruses can control microbial species diversity. Proc Natl Acad Sci USA. 2014;111:7813–8.
Våge S, Pree B, Thingstad TF. Linking internal and external bacterial community control gives mechanistic framework for pelagic virus-to-bacteria ratios. Environ Microbiol. 2016;18:3932–48.
Weitz JS, Stock CA, Wilhelm SW, Bourouiba L, Coleman ML, Buchan A, et al. A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes. ISME J. 2015;9:1352–64.
Gause GF. Experimental analysis of Vito Volterra's mathematical theory of the struggle for existence. Science. 1934;79:16–7.
Talmy D, Beckett SJ, Zhang AB, Taniguch DAA, Weitz JS, Follows MJ. (2019). Contrasting controls on microzooplankton grazing and viral Infection of microbial prey. Front Mar Sci. 2019;6.
Murray AG, Jackson GA. Viral dynamics: a model of the effects of size, shape motion and abundance of single celled planktonic organisms and other particles. Mar Ecol Prog Ser. 1992;89:103–16.
Blackburn N, Zweifel UL, Hagstrom A. Cycling of marine dissolved organic matter .2. A model analysis. Aquat Micro Ecol. 1996;11:79–90.
Våge S, Storesund JE, Giske J, Thingstad TF. Optimal defense strategies in an Idealized microbial food web under trade-off between competition and defense. PloS ONE. 2014;9:e101415.
Abrams PA. Modelling the adaptive dynamics of traits involved in inter- and intraspecific interactions: An assessment of three methods. Ecol Lett. 2001;4:166–75.
Edwards KF, Kremer CT, Miller ET, Osmond MM, Litchman E, Klausmeier CA. Evolutionarily stable communities: a framework for understanding the role of trait evolution in the maintenance of diversity. Ecol Lett. 2018;21:1853–68.
Sandaa R-A, Pree B, Larsen A, Vage S, Topper B, Topper JP, et al. The response of heterotrophic prokaryote and viral communities to labile organic carbon inputs is controlled by the predator food chain structure. Viruses. 2017;9:E238.
Hessen D, VanDonk E. Morphological changes in Scenedesmus induced by substances released from Daphnia. ArcHydrobiol. 1993;127:129–40.
Matz C, Jürgens K. Interaction of nutrient limitation and protozoan grazing determines the phenotypic structure of a bacterial community. Microb Ecol. 2003;45:384–98.
Salcher MM, Pernthaler J, Psenner R, Posch T. Succession of bacterial grazing defense mechanisms against protistan predators in an experimental microbial community. Aquat Micro Ecol. 2005;38:215–29.
Franze G, Pierson JJ, Stoecker DK, Lavrentyev PJ. Diatom-produced allelochemicals trigger trophic cascades in the planktonic food web. Limnol Oceanogr. 2018;63:1093–108.
Thingstad TF, Øvreås L, Egge JK, Løvdal T, Heldal M. Use of non-limiting substrates to increase size; a generic strategy to simultaneously optimize uptake and minimize predation in pelagic osmotrophs? Ecol Lett. 2005;8:675–82.
Tsagaraki TM, Pree B, Leiknes O, Larsen A, Bratbak G, Ovreas L, et al. Bacterial community composition responds to changes in copepod abundance and alters ecosystem function in an Arctic mesocosm study. ISME J. 2018;12:2694–705.
Winter C, Bouvier T, Weinbauer MG, Thingstad TF. Trade-offs between competition and defense specialists among unicellular planktonic organisms: The “Killing the Winner” hypothesis revisited. Microbiol Mol Biol Rev. 2010;74:42–57.
Weitz JS, Hartman H, Levin SA. Coevolutionary arms races between bacteria and bacteriophage. Proc Natl Acad Sci USA. 2005;102:9535–40.
Menge DNL, Weitz JS. Dangerous nutrients: evolution of phytoplankton resource uptake subject to virus attack. J Theoret Biol. 2009;257:104–15.
Litchman E, Klausmeier CA, Schofield OM, Falkowski PG. The role of functional traits and trade-offs in structuring phytoplankton communities: scaling from cellular to ecosystem level. Ecol Lett. 2007;10:1170–81.
Ferenci T. Trade-off mechanisms shaping the diversity of bacteria. Trends Microbiol. 2016;24:209–23.
Abedon ST, Hyman P, Thomas C. Experimental examination of bacteriophage latent-period evolution as a response to bacterial availability. Appl Environ Microbiol. 2003;69:7499–506.
Goldhill DH, Turner PE. The evolution of life history trade-offs in viruses. Curr Opin Virol. 2014;8:79–84.
Parikka KJ, Le Romancer M, Wauters N, Jacquet S. Deciphering the virus-to-prokaryote ratio (VPR): insights into virus-host relationships in a variety of ecosystems. Biol Rev. 2017;92:1081–1100.
Wigington CH, Sonderegger D, Brussaard CPD, Buchan A, Finke JF, Fuhrman JA, et al. Re-examination of the relationship between marine virus and microbial cell abundances (vol 1, pg 15024, 2016). Nat Microbiol. 2017;2:1571–1571.
Parvinen K, Heino M, Dieckmann U. Function-valued adaptive dynamics and optimal control theory. J Math Biol. 2013;67:509–33.
Våge S, Bratbak G, Egge J, Heldal M, Larsen A, Norland S, et al. Simple models combining competition, defence and resource availability have broad implications in pelagic microbial food webs. Ecol Lett. 2018;21:1440–52.
Dugdale RC, Goering JJ. Uptake of new and regenerated forms of nitrogen in primary productivity. Limnol Oceanogr. 1967;12:196–206.
Acknowledgements
This work was supported by EU H2020-INFRAIA project Aquacosm (No 731065), Trond Mohn Stiftelse starting grant SIMPLEX (TMS2019REK02) and RCN project VirVar (No 294363). The authors are grateful for constructive comments by the editor and three anonymous reviewers.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Thingstad, T.F., Våge, S. Host–virus–predator coexistence in a grey-box model with dynamic optimization of host fitness. ISME J 13, 3102–3111 (2019). https://doi.org/10.1038/s41396-019-0496-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-019-0496-7