Abstract
Significant gaps in our understanding of how global change drivers interact to affect the resistance and functioning of microbial communities hinders our ability to model ecosystem responses and feedbacks to co-occurring global stressors. Here, we investigated the effects of extreme drought and exotic plants, two of the most significant threats to Mediterranean-type ecosystems, on soil microbial community composition and carbon metabolic genes within a four-year field rainfall manipulation experiment. We combined measurements of bulk microbial and soil properties with high-throughput microbial community analyses to elucidate microbial responses and microbial-mediated alterations to carbon cycling. While microbial responses to experimental droughts were weak, scant rainfall periods resulted in decreased microbial biomass and activity, and relative abundances of bacterial groups such as Proteobacteria, Verrucomicrobia, and Acidobacteria decreased concomitantly with increases in Actinobacteria, Chloroflexi, and Firmicutes abundance. Soils under exotic plants had increased temperatures, enhanced infiltration during rainfall events, and decreased water retention and labile carbon in comparison to soils under native plants. Higher peaks and more seasonally variable microbial activity were found under exotic plants and, like drought periods, the microbial community shifted towards osmotic stress life-strategies. Relationships found between microbial taxonomic groups and carbon metabolic genes support the interpretation that exotic plants change microbial carbon cycling by altering the soil microclimate and supplying easily decomposed high-quality litter. Soil microbial community responses to drought and exotic plants could potentially impact ecosystem C storage by producing a smaller, more vulnerable C pool of microbial biomass that is prone to increased pulses of heterotrophic respiration.
Similar content being viewed by others
Introduction
Exotic plants and climate disturbances are among the most significant threats to Mediterranean-type ecosystems (MTEs) [1, 2]. This is the case for shrub-dominated MTEs distributed along coastal and southern California (CA) that have been invaded extensively by exotic forbs and grasses [3] and have recently experienced 5 consecutive years (2012–2016) of high temperatures and scant rainfall, including three consecutive years of record warmth (2014–2016) (NOAA-NCEI). With projected future scenarios of increased drought severity [4], there is a need to improve our understanding of the interactions of these co-occurring stressors and potential consequences for ecosystem functions. Losses in biodiversity and carbon (C) storage are of particular concern as MTEs have been underscored as global reservoirs of biodiversity [5] and play an important role in the interannual variability of global C sequestration [6].
Numerous studies of the effects of exotic plants and rainfall shifts on CA shrublands have had an aboveground focus, highlighting the role of the competitive advantages of exotic plants [7,8,9,10,11] and soil nitrogen feedbacks in ecosystem alterations [12, 13]. However, fewer studies have considered belowground communities [13, 14] and impacts on soil C cycling [15]. While often overlooked, soil communities represent one of the largest reservoirs of biodiversity [16] and comprise a significant fraction of soil carbon [17,18,19,20]. Additionally, microbial heterotrophic respiration is one of the largest avenues of C loss from ecosystems [21]. Thus, assessments of the response of MTE soil communities to global stressors can enhance our understanding of community dynamics and how these responses influence ecosystem C functions [22].
A growing body of belowground-based studies have reported the resistance and resilience of semi-arid soil microbial communities to extreme weather events due to their routine exposure to seasonal drought and ability to recover upon rewetting [23,24,25,26,27,28]. While mounting evidence suggests that changes in community structure can occur as adaptive responses to drying-rewetting cycles [29, 30], responses by microbial communities to multiple global change disturbances remain poorly understood [31].
Exotic plants can interact with climatic changes to affect soil microbial communities by physically altering soil microclimates and by altering plant-microbe interactions [22, 32, 33]. Exotic species common to CA MTEs have been reported to have shallower roots [34, 35], shorter life spans, earlier seasonal activity [11], and higher litter quantity and quality [15] when compared to dominant native shrubs. The differentiation in plant structures as well as the timing, quantity, and quality of C sources between CA MTEs native and exotic plants can alter microbial communities and hence their responses to extreme droughts [36].
In MTEs, microbial responses to co-occurring global stressors must be considered in light of temporal variation, as intra-annual and inter-annual variability are key drivers of ecosystem processes [37]. As shown in previous studies that examined the response of microbial communities over multiple temporal scales [23,24,25, 27], there is a need to gather experimental evidence that incorporates temporally explicit assessments. Furthermore, recent high-throughput DNA sequencing technology presents an opportunity to characterize and quantify soil microbial functional attributes in greater depth, providing insight into microbial resistance [38] and ecosystem functions [39].
Here, we monitored soil microbial biomass, activity, community composition, and C functional genes under native-dominated and exotic-dominated, rainfall manipulated plots in a CA MTE community experiencing a multi-year drought to assess the impacts of extreme drought, exotic plants and temporal variability on soil microbial communities and carbon cycling. We hypothesized that physiological stress caused by extreme drought conditions could impact microbial population size, shift microbial community structure, and alter microbial functions. Alternatively, while microbial communities are generally constrained by droughts, the adaptive capabilities and phenotypic plasticity of soil microbes may yield mechanisms to tolerate and resist periods of droughts. We expected complex interactions between temporal variability, plant community types, and microbial communities. For example, the lack of aboveground structures in exotic annuals can exacerbate the impacts of a drying climate to promote a more stressful environment that is detrimental for soil microbes [40]. On the other hand, higher quality exotic litter can promote microbial abundance and metabolism, altering microbial C processes [41].
Materials and methods
Site description
This study was conducted at the Santa Margarita Ecological Reserve (SMER), located in the upper part of the Santa Margarita River watershed (33˚26′28.6″N, 117˚09′ˊ51.89″W), in Riverside County, CA, USA. The site is dominated by native mature shrubs, such as Artemisia californica, Salvia mellifera, and Salvia apiana, interspersed with native perennial herbs including Calochortus splendens and Dichelostemma capitatum and exotic annual species, such as Centaurea melitensis, Hirschfeldia incana, and Bromus madritensis. These exotic species likely became established after the site was disturbed in the 1970s during the construction of a water pipeline. The region has a 30-year (1981–2010) rainfall average of 358 mm year−1.
Thirty 3 m × 3 m experimental plots with a dominant plant community composition of either native shrub (n = 15) or exotic annuals (n = 15) were established prior to the 2013 rainy season. Salvia mellifera and Artemisia californica dominated the native plots and exotic annuals, such as Centaurea melitensis, Hirschfeldia incana and Bromus madritensis, dominated the exotic plots. All plots were located on similar soil, classified as Las Posas loam, Fine, smectitic, thermic Typic Rhodoxeralfs, which are well drained with an argilic horizon under topsoil that ranges from 15 to 46 cm (http://websoilsurvey.sc.egov.usda.gov). Soil analyses at the beginning of the experiment showed no differences in pH, soil organic matter, and texture between plots (Supplementary Fig. 1), indicating that these parameters, the major variables responsible for shaping soil microbial communities [42,43,44], were consistent among treatments. We are therefore reasonably confident that the effects observed in this experiment did not arise from unintentional historical artifacts. Using rainfall shelters, three rainfall treatments (50, 100, and 150% of ambient rainfall) were imposed on experimental plots of native vs. exotic plant communities. For each combination of rainfall and plant community type, five replicates were allocated in a randomized block design. The rainfall exclusion shelters were constructed with clear corrugated acrylic roofing and were removed from May to September each year (dry season). Rainfall was collected from the roofs by gutters and stored in on-site tanks for redistribution to plots to achieve each respective rainfall treatment level using an overhead irrigation system. The irrigation system was controlled by volume flow meters. Daily rainfall was monitored from the NOAA Fallbrook 5 NE weather station (http://www.ncdc.noaa.gov/qclcd/QCLCD?prior=N) located 2.4 km east of the study site. After each rain event, the appropriate amount of watering was applied to plots to achieve the rainfall treatments.
Soil sampling and analyses
To evaluate how intra-annual and inter-annual variations influence belowground microbial dynamics, soils were sampled during the winter (rainy season), spring (peak plant biomass), and summer (dry season) over a 4-year period from 2013 to 2016, resulting in 12 sampling events (Supplementary Table 1). Soil was sampled to a depth of approximately 15 cm using a 5 × 15 cm slide hammer soil core sampler (ASM Inc.). Samples were stored in a cooler for transportation and processed within 24 h of collection. All visible stones, roots and litter were removed with forceps. To assess whether rainfall treatments and plant community type influenced the soil microclimate, we measured the gravimetric water content (GWC) of soil samples by drying 5-g of soil at 65 °C for 24 h and measured soil temperature in the field during 4 of the 12 sampling times (Summer 2013 and 2015 and Winter and Spring of 2014) using the temperature sensor of an EGM-4 Environmental Gas Monitor (PP Systems). To evaluate alterations in soil C, dissolved organic carbon (DOC) was measured in 0.5 M potassium sulfate extracts, determined by oxidation with Mn3+, and estimated on a spectrophotometer [45].
Microbial community analyses
To test for alterations in microbial population size, microbial biomass carbon (MBC) was evaluated by the fumigation-extraction (FE) method [46]. After extractions (in 0.5 M potassium sulfate), the C concentrations in the extracts were determined [45]. The MBC contents were calculated from the difference between fumigated and non-fimigated samples. Substrate-induced respiration (SIR) (Anderson and Domsch 1978) was used to assess microbial activity as SIR is a simple and rapid method that provides a suitable index of potentially active biomass [47]. Bacterial community structure and C functional genes analyses were performed on a subsample of soils (n = 37) collected in the spring months of 2013 (4 native and 4 exotic), 2015 (6 native and 5 exotic) and 2016 (9 native and 9 exotic) (Supplementary Table 2). Soil DNA was extracted using Power Soil DNA isolation kit (MoBio, Carlsbad, CA USA) according to the manufacturer’s instructions. Extracted DNA was quantified by PicoGreen fluorometry and diluted with sterile water to 30 ng µL−1 DNA based on the lowest sample concentration. The diluted DNA was sheared to 350 base pairs (bp) using a Covaris ultrasonicator (Covaris Inc., Woburn, MA, USA) according to manufacturer’s recommendations. The sheared DNA was repaired, ligated, and indexed using the Accel-NGS 2S Plus DNA Library Kit (Swift Biosciences, Inc. Ann Arbor, MI, USA). The prepared library was quantified using Agilent 2100 Bioanalyzer System and sequenced using an Illumina MiSeq (300 bp). Quality control and preprocessing of metagenomic data was performed using PRINSEQ to eliminate sequences with less than 75 bps, less than 25 mean quality scores, and replicated reads [48]. The sequences in each taxon or function were calculated and normalized by using Metagenomic rapid annotations (MG-RAST Version 4) against RefSeq Subsystems databases with an e-value cutoff of 10−5, a minimum identity cutoff of 60%, and a minimum alignment length cuttoff of 15 aa [49].
Statistical analyses
Data analysis was perfomed in R version 3.3.3 [50]. To determine the effects of rainfall treatment, plant community type, season, and year and their interactions on MBC, SIR, GWC, and DOC, mixed-effects repeated measures analysis of variance (ANOVA) was employed using the lme function within the nlme package. Linear model assumptions of normality and homoscedasticity were tested using Shapiro-Wilk and Levene’s test and observed visually using Q–Q plots and plotting residuals against fitted values, respectively. When significant effects were found, multiple comparisons between treatments, seasons, and years were performed using the glht function within the multcomp package. To test significant correlations, Pearson correlation tests were performed using cor.test function. P-values less than 0.05 were considered significant.
To determine the effects of plant community type, rainfall treatment, and interannual variability on bacterial community structure and carbon cycling genes, taxonomic profiles were compared at the phylum, class and genus levels, and functional profiles were compared at the level 2 subsystems that belonged to carbohydrate metabolism [51]. Bray–Curtis dissimilarities were compared by permunational multivariate analysis of variance (PerMANOVA) using the adonis function of the vegan package [52]. When significant effects were found, multiple comparisons between treatments and years were performed using the pairwise.perm.manova function within the RVAideMemoire package. Non-metric multidimensional scaling (NMDS) analysis was also used to visualize community similarity between metagenomes [53] using the function ‘metaMDS’ in the vegan package. Component scores were used to generate 95% confidence ellipses around metagenomes using the ordiellipse() function within vegan. Also within vegan, a Pearson’s correlation matrix was constructed for percent abundances of gene hits corresponding to most abundant phyla and abundances of level 2 carbohydrate metabolism using as .matrix() and cor() functions [54]. Statistical analysis of the Metagenomic Profiles (STAMP) (version v2.0.3) was also used to identify statistically significant taxonomic and functional groups among year, rainfall treatments, and plant community type [55]
Results
Effects of droughts and exotic plants on soil microclimate
During the 42-month sampling period (December 2012–June 2016), CA was under natural drought conditions. Our field site received 209, 218, 285, and 207 mm of rainfall during the water years of 2013–2016, respectively, compared to a long-term average of 358 mm year−1 (Table 1). Additionally, a dry year also occurred the year before the experiment (2012), when the site received 285 mm of rainfall. Due to the lack of rainfall, our 50 and 100% rainfall treatments represented two levels of drought (mean = 32 and 64% of normal rainfall, respectively) and the 150% treatment corresponded to a normal rainfall year. Water year 2014 had the driest winter, receiving only 122 mm from October to February (Table 1). Water year 2015 was the wettest year during the study, receiving major Pacific storms and summer rains associated with the 2015–2016 El Niño event.
Rainfall treatment effects on surface (0–10 cm) gravimetric soil moisture were observed more strongly during the winter and spring seasons than during summer dry seasons, when GWC values fell to similar levels for all treatments (rainfall treatment × season interaction P = 0.002, Table 2). For example, during the winters GWC values in native-dominated plots averaged 9, 11, and 12% (based on average values across all winters) in the 50, 100, and 150% rainfall treatments and 8, 12, and 14% in the exotic-dominated plots. During the summers GWC dropped to approximately 3% for all rainfall treatments in both plant community types (Fig. 1a). Post-hoc comparisons detected differences between the 50 and 150% treatments especially during the winter seasons of 2015 and during the spring seasons of 2013 and 2014 (rainfall treatment × season × year interaction, P = 0.04), potentially associated with high rainfall events (Table 1).
Plant community type influenced soil microclimate through direct and interactive effects. The effects of plant community on soil moisture varied by season and year (plant × season × year interaction P = 0.002). During the first year (2013) higher levels of GWC were found during the winter and summer in native-dominated plots when seasons were evaluated separately (Fig. 1a). As drought duration increased, higher levels of GWC were observed in exotic-dominated plots throughout the winter seasons (although only significant in 2015). During the springs, significantly higher GWC values were found in native-dominated plots in 2014 and in exotic dominated plots in 2016. Plant community composition also affected soil temperature significantly. On average, a 3-degree difference was detected between plant community types (estimated mean ± SD, native-dominated: 20.3 ± 6.5 vs. exotic-dominated: 23.3 ± 7.6 °C, based on average values across all observations).
Microbial population size and activity responses
Microbial bulk properties (MBC, SIR) responded more strongly to temporal variability than to rainfall treatments or plant community type (Table 2). Given the complex interactions among plant type, season and year, direct treatment effects were weak, but were seen more clearly when years were evaluated separately. For example, the overall effect of plant type on MBC was marginal (P = 0.066), but during the first two years of the experiment (2013–2014), MBC was significantly higher in native-dominated plots (P = 0.03 when 2013 and 2014 were evaluated separately) (Fig. 1b). During 2015 and 2016 no significant differences were found between plant community types (P = 0.77 when 2015 and 2016 were evaluated separately). Similarly, the overall rainfall treatment effect on MBC was weak (P = 0.049), but a highly significant increase in MBC in the 150% treatment was found during 2015 (the wettest year) when compared to the other rainfall treatments (Supplementary Fig. 2).
The highly significant seasonal effect on MBC (P = 0.0002) reflects a trend towards highest biomass in winter and lowest in summer, though these patterns varied by year (Fig. 1b). During 2016, low MBC values were observed during the winter and summer seasons (Fig. 1b). The lack of the 2016 winter pulse could have been associated with the scant rainfall during that winter (Table 1).
The highly significant effect of year on MBC (P < 0.0001) reflects a trend of decreasing biomass over the course of the experiment. This is seen most clearly during the winter seasons when average values of MBC significantly decreased from 2013 to 2014 (in native-dominated plots) and from 2015 to 2016 (in both plant communities) (Supplementary Fig. 3). This pattern correlates with annual rainfall decreasing in consecutive years (Table 1). For both plant communities, a weak but significant correlation was found between MBC and GWC (native-dominated: P = 0.0002 and exotic-dominated: P = 0.004, Fig. 2a).
SIR, a measure of potentially active microbial biomass, was not significantly affected by rainfall treatment or plant type, but showed a similar long-term decline to that seen for MBC. Mean annual SIR decreased over time (2013 > 2014 = 2016 > 2015 in native-dominated plots and 2013 > 2014 = 2015 > 2016 in exotic-dominated plots, Supplementary Fig. 4). Soils in exotic-dominated plots had more pronounced SIR seasonal variation, with higher spikes during spring (2013 and 2014) or summer (2015) (plant × season interaction P = 0.044, Fig. 1c). In native plots, SIR was positively correlated with MBC (native-dominated: P = 0.002 and exotic-dominated: P = 0.2, Fig. 2b) and DOC (native-dominated: P = 0.01 and exotic-dominated: P = 0.3, Fig. 2c).
Bacterial community composition responses
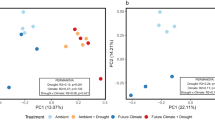
The soil bacterial community, as revealed by metagenomic sequencing, was most strongly influenced by annual variability, but effects of plant community and experimental rainfall treatment were also observed (Table 2). A total of 27 bacterial phyla were detected, with Proteobacteria (36%), Actinobacteria (31%), Bacteroidetes (6%), Firmicutes (6%), Acidobacteria (4%), Chloroflexi (4%), and Verrucomicrobia (3%) accounting for 90% of the sequences. The PerMANOVA results from phylum relative abundances detected significant differences between years and plant community types but not between rainfall treatments (Table 2). Consistent with the PerMANOVA post-hoc comparisons, NMDS analysis showed differences between 2015 communities when compared to 2013 and 2016 (Fig. 3). STAMP showed that 14 bacterial phyla differed significantly between years. Among the most abundant, Actinobacteria and Deinococcus-Thermus were more abundant in 2013, Acidobacteria, Bacteroidetes, Planctomycetes, Proteobacteria, and Verrucomicrobia in 2015 (wettest year) and Actinobacteria, Chloroflexi and Firmicutes in 2016 (Supplementary Fig. 5). At the class level, alpha-Proteobacteria, beta-Proteobacteria, delta-Proteobacteria, and gamma-Proteobacteria, Acidobacteria, Solibacteres, Spartobacteria, and Opitutae were overrepresented in 2015, while Bacilli (Firmicutes) and Dehalococcoidetes (Chloroflexi) were overrepresented in 2016.
The plant community significantly influenced microbial community structure; a significant reduction of Proteobacteria and Verrucomicrobia and an increase in the relative abundance of Chloroflexi, Cyanobacteria and Deinococcus-Thermus were found in exotic-dominated plots (Supplementary Fig. 6). Among these phyla, 84 genera were significantly over- or under-represented in native-dominated and exotic-dominated plots, with 58 belonging to the phylum Proteobacteria, 14 to Cyanobacteria, 5 to Chloroflexi, 4 to Verrucomicrobia, and 3 to Deinococcus-Thermus (Supplementary Fig. 7). The PerMANOVA results at the genus level detected significant differences between rainfall treatments (Table 2). The NMDS analyses shows this effect is driven by the 150% treatment clustering apart from the 50 and 100% treatments (Supplementary Fig. 8).
C cycling
Soil DOC was mostly influenced by plant community type (P = 0.0002) and season and year interactions (P < 0.0001) (Table 2). DOC was reduced in exotic plots by approximately 25% based on average values of all plots across the entire time of observations (estimated mean ± SD, native-dominated: 61.3 ± 35.6 vs. exotic-dominated: 45.8 ± 23.8 μg C g−1 soil). Seasonal effects were detected after post-hoc comparisons during 2013 and 2014, when DOC peaked during the winter season, and in 2015 during the summer season for both plant community types (Fig. 1d).
Carbon metabolism represented 15% of the broad scale (level 1) bacterial functional genes with a total of 11 functional attributes at the level 2 (Fig. 4). PerMANOVA results detected significant differences between years and plant community types but not between rainfall treatments (Table 2). Contrary to the annual trends for taxonomy, in which 2015 was distinct from the other years, post-hoc comparisons and NMDS analysis of functional gene abundance showed differences between 2013 vs. 2015 and 2016 (Fig. 3).
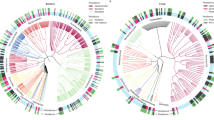
Pearson’s correlation matrix between carbon cycling functional gene abundances (x-axis) and microbial phyla abundances (y-axis). Functional genes were clustered by Bray–Curtis dissimilarity and Ward’s distance, and groupings informally annotated. Pearson’s correlation is color coded with blue being positive and red being negatively correlated. Summary statistics (boxplot) for each row are provided
There was a striking commonality between the impacts of wet vs. dry years and the influence of native vs. exotic plant communities on the microbial community structure. Hence, the annual and vegetation-driven variations in microbial community were generalized into a single axis with two groups, with group 1 representing exotic plots and the driest years (2013 and 2016) and group 2 representing native plots and the most mesic year (2015) (Fig. 4). Correlation analyses of these groups and C cycling genes showed two distinct taxonomic/functional clusters (Fig. 4). Group 1, composed of bacterial groups Actinobacteria, Chloroflexi, Cyanobacteria, Deinococcus-Thermus, and Firmicutes, were positively correlated with aminosugars and sugar alcohols, and Group 2, composed of Acidobacteria, Bacteroidetes, Planctomycetes, Proteobacteria, and Verrucomicrobia, were mostly positive correlated with polysaccharides and glycoside hydrolases (Fig. 4).
Discussion
The results of this experimental study showed that: (1) microbial population size, structure, and functioning responded more to temporal rainfall variability than experimental droughts, (2) in comparison to soils under native plants, soils under exotic plants differed in microbial activity, structure, and functioning through complex interactions, and (3) microbial responses to climate variability and exotic plants can influence soil carbon cycling processes.
Recent evidence suggests that extreme seasonal changes to Mediterranean-type ecosystems may have a larger impact on soil communities than observed in experimental droughts [24, 39]. In this study, we found that microbial communities were more responsive to temporal variability than to experimental droughts. However, we observed responses of microbial communities to multi-year droughts and rainfall shifts. For example, we observed microbial biomass and activity declining over the extended (4-year) period of drought, with significant declines during summer seasons, with the exception of 2015, which experienced high summer rainfall. During 2015 (the wettest year), microbial biomass significantly increased in the 150% treatment when compared to the other rainfall treatments. Other rainfall manipulation studies have also shown a positive response in microbial characteristics with high rainfall [25, 56]. Zhao et al. [56] detected an increment in microbial biomass with increased rainfall up to 40%+ of ambient rainfall in a semi-arid steppe, and Cruz-Martínez et al. [25] detected microbial community differences between rainfall treatments when the watering event exceeded normal conditions. Water is clearly a strong factor controlling microbial activity and diversity, but its effect is strongly dependent on timing. While MBC was correlated with GWC, this relationship explained only a small amount of variation (Fig. 2a). The relatively large influence of seasonal and annual variation in rainfall compared to the effects of individual rainfall event size and soil water content suggest that the timing of rainfall events can be more important than rain intensity [57]. The annual effects could also arise from variations in temperature, humidity and their resulting impacts on the plant community. The exotic plant community in this ecosystem is particularly dynamic, varying annually in relative dominance of grasses and forbs (E.E.C., unpublished data).
Due to sampling frequency and the methods employed, our experimental design did not address immediate, short-term responses of microbes to soil drying and rewetting. While FE methods can include inactive microbes, the detection of seasonal and annual changes in MBC (Fig. 1 and Supplementary Fig. 3) suggests that FE measurements capture microbial turnover at seasonal and annual time scales. Correlations between MBC and the physiologically active biomass (SIR) also support this conclusion.
Bacterial community composition behaved similarly to microbial biomass, responding more strongly to year than to rainfall treatments, despite the possibility that persistence of relic DNA may have limited our ability to detect changes over time. Higher abundance of gram-positive bacteria, such as Actinobacteria, Chloroflexi and Firmicutes, found during 2013 and 2016 (drier years) vs. gram-negative groups, such as Proteobacteria, Verrucomicrobia, and Acidobacteria, that were found in higher abundance in 2015 (wetter year) may indicate that the composition shifted due to osmotic stress. Gram-positive bacteria are known to be more drought tolerant (stronger cell wall and drought avoidance strategies such as spore formation) than gram-negative bacteria [58]; thus we speculate that these microbial taxonomic shifts were likely due to osmotic stress caused by low rainfall during 2013 and 2016. This is consistent with Castro et al. [59], Cruz-Martínez et al. [60], and Neilson et al. [61] which found similar relationships between moisture and bacterial communities. However, Castro et al. [59] found Acidobacteria were more abundant under dry conditions which has been also found in a long-term drought treatment [39]. Variance in the responses of Acidobacterial groups has been explained in terms of distinct classes within this phyla [62], and consistent with our finding, the group Solibacteres has been found to decrease with drying in a summer desiccation experiment [63]. Additionally, while this study observed compositional changes among years, we observed several phyla-level responses to annual rainfall variability that are also consistent with drying and rewetting studies [26, 30]. For example, Barnard et al. [30] found Actinobacteria and Firmicutes to decrease while Acidobacteria and Verrucobacteria increased during soil rewetting, a pattern that we found during our wettest year. Similar to our findings, Drigo et al. [26] found Deinococcus taxa resistant to drought conditions.
Our results showed that soils under exotic plants had increased temperatures, enhanced infiltration during rainfall events, decreased water retention during spring and summer, and decreased labile carbon in comparison to soils under native plants. Similar impacts have been reported in shrubs vs. grasses studies, where increases in runoff and evaporation due to the lack of soil retention capacity and shading conditions were found in grass-dominated systems [64,65,66]. We propose that these soil hydraulic alterations under exotic plants represent an amplification of the rewetting-drying cycles of MTEs and result in a more heat and osmotically stressed environment. Reductions in microbial biomass, enhanced seasonal fluctuations in MBC and SIR, and reductions in less tolerant bacterial groups such as Proteobacteria and Verrucomicrobia in exotic dominated plots support this theory. Overlaps in taxonomic and functional composition between exotic plots and 2016 (hotter and 5th consecutive drought year) and between native plots and 2015 (wetter year) (Fig. 3) further support our interpretation that soils under exotic plants stimulate osmotic and heat stresses. Additionally, increases of di- and oligosaccharide metabolic genes during 2016 and in exotic plots can be attributed to osmotic stress adaptations since disaccharides, such as trehalose, have been previously reported as compatible solutes in osmoregulation [67, 68].
While harsher environmental conditions under exotic plants may negatively impact microbial communities, it is also possible that the higher temperatures and rainfall water pulses coupled with the more easily decomposed, high quality litter of exotics may enhance microbial respiration. The higher spikes and more pronounced microbial seasonal activity (Fig. 1c) found in this experiment can be supported by the enhancement of soil microbial respiration with increasing temperature [69] and findings of higher soil respiration with additions of exotic plant litter in CA MTEs when compared to native litter, especially under high moisture pulse scenarios [70]. These findings help explain microbial mechanisms for the reduction of soil C in invaded MTEs [71, 72]. By increasing soil temperature, water pulses, and litter quality, exotic plants can promote microbial respiration and hence, microbial-mediated C losses. Additionally, while not explicitly tested in this study, increases in soil temperature found in exotic plots also support findings of ecosystem C loss via photodegradation [73]. This effect of exotic plants was also reflected in the significant reductions of DOC in exotic dominated plots (Fig. 1d). While DOC represents only a fraction of soil C, labile C fractions have been suggested as an appropriate metric for short-term (<15 years) changes of soil C pools in semi-arid ecosystems since bulk soil C can respond relatively slowly [74].
In addition to microclimatic effects and litter quality, there are a host of other traits that differ between native shrubs and exotic herbs, such as rooting depth, C allocation [34] and plant performance under stressful conditions [75,76,77] that could have contributed to the observed effects of plant type. It is also possible that some residual effect of the disturbance that led to invasion of this landscape influenced our results. However, such effects are probably minimal, given the similarity of soil physical factors between the two treatments and the fact that the establishment of both native and exotic plants in this ecosystem are generally limited by dispersal of seeds to bare ground [78, 79] rather than special soil requirements.
When taken together, our results showed two distinct taxonomic/functional groups that can be associated with life-history traits, specific C sources, and microbial C uses. Group 1, composed of Actinobacteria, Chloroflexi, Cyanobacteria, Firmicutes and Deinococcus-Thermus (considered extremophiles), were better represented under exotic plants and were positively correlated with genes for aminosugars, sugar alcohols and simple carbohydrate metabolic pathways, such as di- and oligosaccharides, which include many compatible solutes that confer stress tolerance. These correlations indicate that Group 1 microbes rely on recycling microbial biomass (amino sugars in cell walls) and C forms released during rainfall events (compatible solutes), especially after enhanced drought conditions [80]. Group 2, composed by bacterial groups such as Verrucomicrobia and Acidobacteria, which are generally considered K-selected [81], were better represented under native plants and correlated with complex carbohydrate metabolic genes, such as polysaccharides and glycoside hydrolases, which are included in plant structures [82,83,84] (Fig. 4). This indicates that Group 2 microbes rely more on plant litter and root exudates [82, 83]. This is consistent with findings of Group 1 member Verrucomicrobia correlating with polysaccharide metabolism [85].
The relationships found in this experiment between microbial community structure, functional genes and environmental factors are not strictly reflective of copiotrophic/r-selected and oligotrophic/K-selected taxa generalizations [81, 86]. A possible explanation is that more extreme pulses of rain penetration and microbial activity in exotic-dominated soils result in undetected, short-lived carbon rich conditions that are rapidly metabolized by microbes. Additionally, the more recalcitrant native litter may select for a microbial community adapted to complex carbon compounds, allowing for the accumulation of DOC, as supported by the positive relationship between Group 2 and complex carbohydrate metabolism. Other explanations for low DOC in exotic dominated plots are leaching and runoff losses promoted by the higher rainfall water pulses.
Conclusions
In this MTE, we found that temporal variability and plant traits are key factors in assessing microbial responses to extreme droughts and exotic plants disturbances. We observed extreme periods of scant rainfall to decrease microbial biomass and activity and alter microbial communities. In comparison to soils under native plants, soils under exotic plants characteristically had a microbial environment prone to increased osmotic and heat stress that enhanced microbial turnover and loss of labile soil C. Correlations of microbial taxa and C metabolic profiles in response to climate and plant disturbances suggested relationships between specific microbial traits and potential C processes. Soil microbial community responses to extreme droughts and exotic plants could potentially impact ecosystem C storage by producing a smaller, more vulnerable microbial biomass C pool prone to increased pulses of heterotrophic respiration. While this study observed microbial responses to multi-year droughts, future studies are needed to understand the resilience of microbial communities after periods of extreme droughts and potential implications for carbon cycling [87].
References
Chapin FS, Zavaleta ES, Eviner VT, Naylor RL, Vitousek PM, Reynolds HL, et al. Consequences of changing biodiversity. Nature. 2000;405:234–42.
Sala OE, Chapin FS, Armesto JJ, Berlow E, Bloomfield J, Dirzo R, et al. Global biodiversity scenarios for the year 2100. Science. 2000;287:1770–4.
Minnich RA, Dezzani RJ. Historical decline of coastal sage scrub in the Riverside-Perris Plain, California. West Birds. 1998;29:366–91.
Dai A. Increasing drought under global warming in observations and models. Nat Clim Change. 2012;3:52–8.
Myers N, Mittermeier RA, Mittermeier CG, Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–8.
Poulter B, Frank D, Ciais P, Myneni RB, Andela N, Bi J, et al. Contribution of semi-arid ecosystems to interannual variability of the global carbon cycle. Nature. 2014;509:600–3.
Ashbacher AC, Cleland EE. Native and exotic plant species show differential growth but similar functional trait responses to experimental rainfall. Ecosphere. 2015;6:1–14.
Cleland EE, Esch EH, McKinney J. Priority effects vary with species identity and origin in an experiment varying the timing of seed arrival. Oikos. 2015;124:33–40.
Goldstein LJ, Suding KN. Intra-annual rainfall regime shifts competitive interactions between coastal sage scrub and invasive grasses. Ecology. 2014;95:425–35.
Wainwright CE, Cleland EE. Exotic species display greater germination plasticity and higher germination rates than native species across multiple cues. Biol Invasions. 2013;15:2253–64.
Wainwright CE, Wolkovich EM, Cleland EE. Seasonal priority effects: implications for invasion and restoration in a semi-arid system. J Appl Ecol. 2012;49:234–41.
Dickens SJM, Allen EB. Soil nitrogen cycling is resilient to invasive annuals following restoration of coastal sage scrub. J Arid Environ. 2014;110:12–8.
Mauritz M, Cleland EE, Merkley M, Lipson DA. The influence of altered rainfall regimes on early season N partitioning among early phenology annual plants, a late phenology shrub, and microbes in a semi-arid ecosystem. Ecosystems. 2014;17:1354–70.
Dickens SJM, Allen EB, Santiago LS, Crowley D. Exotic annuals reduce soil heterogeneity in coastal sage scrub soil chemical and biological characteristics. Soil Biol Biochem. 2013;58:70–81.
Wolkovich EM, Lipson DA, Virginia RA, Cottingham KL, Bolger DT. Grass invasion causes rapid increases in ecosystem carbon and nitrogen storage in a semiarid shrubland. Glob Change Biol. 2010;16:1351–65.
Bardgett RD, Van Der Putten WH. Belowground biodiversity and ecosystem functioning. Nature. 2014;505:505–11.
Joergensen RG, Wichern F. Alive and kicking: why dormant soil microorganisms matter. Soil Biol Biochem. 2018;116:419–30.
Kallenbach CM, Grandy AS, Frey SD, Diefendorf AF. Microbial physiology and necromass regulate agricultural soil carbon accumulation. Soil Biol Biochem. 2015;91:279–90.
Khan KS, Mack R, Castillo X, Kaiser M, Joergensen RG. Microbial biomass, fungal and bacterial residues, and their relationships to the soil organic matter C/N/P/S ratios. Geoderma. 2016;271:115–23.
Liu N, Zhang Y, Chang S, Kan H, Lin L. Impact of grazing on soil carbon and microbial biomass in typical steppe and desert steppe of inner mongolia. PLoS One. 2012. https://doi.org/10.1371/journal.pone.0036434.
Chapin FS, Matson PA, Vitousek PM. Principles of terrestrial ecosystem ecology. New York: Springer; 2012.
Classen AT, Sundqvist MK, Henning JA, Newman GS, Moore JAM, Cregger MA, et al. Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: What lies ahead? Ecosphere. 2015;6:art130.
Canarini A, Carrillo Y, Mariotte P, Ingram L, Dijkstra FA. Soil microbial community resistance to drought and links to C stabilization in an Australian grassland. Soil Biol Biochem. 2016;103:171–80.
Cregger MA, Schadt CW, McDowell NG, Pockman WT, Classen AT. Response of the soil microbial community to changes in precipitation in a semiarid ecosystem. Appl Environ Microbiol. 2012;78:8587–94.
Cruz-Martínez K, Suttle KB, Brodie EL, Power ME, Andersen GL, Banfield JF. Despite strong seasonal responses, soil microbial consortia are more resilient to long-term changes in rainfall than overlying grassland. ISME J. 2009;3:738–44.
Drigo B, Nielsen UN, Jeffries TC, Curlevski NJA, Singh BK, Duursma RA, et al. Interactive effects of seasonal drought and elevated atmospheric carbon dioxide concentration on prokaryotic rhizosphere communities. Environ Microbiol. 2017;19:3175–85.
Hueso S, Hernández T, García C. Resistance and resilience of the soil microbial biomass to severe drought in semiarid soils: the importance of organic amendments. Appl Soil Ecol. 2011;50:27–36.
Yuste JC, Peñuelas J, Estiarte M, Garcia-Mas J, Mattana S, Ogaya R, et al. Drought-resistant fungi control soil organic matter decomposition and its response to temperature. Glob Change Biol. 2011;17:1475–86.
Evans SE, Wallenstein MD. Climate change alters ecological strategies of soil bacteria. Ecol Lett. 2014;17:155–64.
Barnard RL, Osborne CA, Firestone MK. Changing precipitation pattern alters soil microbial community response to wet-up under a Mediterranean-type climate. ISME J. 2015;9:946–57.
Caldeira MC, Lecomte X, David TS, Pinto JG, Bugalho MN, Werner C. Synergy of extreme drought and shrub invasion reduce ecosystem functioning and resilience in water-limited climates. Sci Rep. 2015;5:1–9.
Garbeva P, van Veen JA, van Elsas JD. Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol. 2004;42:243–70.
Bardgett RD, Wardle DA. Aboveground-belowground linkages. Biotic interactions, ecosystem processes and global change. Oxford Series in Ecology and Evolution. Oxford, UK: Oxford University Press; 2010.
Koteen LE, Baldocchi DD, Harte J, Koteen LE, Baldocchi DD. Invasion of non-native grasses causes a drop in soil carbon storage in California grasslands. Environ Res Lett. 2011;6:044001.
Wood YA, Meixner T, Shouse PJ, Allen EB. Altered ecohydrologic response drives native shrub loss under conditions of elevated nitrogen deposition. J Environ Qual. 2006;35:76–92.
Lange M, Eisenhauer N, Sierra Ca, Bessler H, Engels C, Griffiths RI, et al. Plant diversity increases soil microbial activity and soil carbon storage. Nat Commun. 2015;6:6707.
Snyder KA, Tartowski SL. Multi-scale temporal variation in water availability: Implications for vegetation dynamics in arid and semi-arid ecosystems. J Arid Environ. 2006;65:219–34.
de Vries FT, Shade A. Controls on soil microbial community stability under climate change. Front Microbiol. 2013;4:265.
Curiel Yuste J, Fernandez-Gonzalez AJ, Fernandez-Lopez M, Ogaya R, Penuelas J, Sardans J, et al. Strong functional stability of soil microbial communities under semiarid Mediterranean conditions and subjected to long-term shifts in baseline precipitation. Soil Biol Biochem. 2014;69:223–33.
Wolf AA, Zavaleta ES, Selmants PC. Flowering phenology shifts in response to biodiversity loss. Proc Natl Acad Sci. 2017;114:3463–8.
Liao C, Peng R, Luo Y, Zhou X, Wu X, Fang C, et al. Altered ecosysetm carbon and nitrogen cycles by plant invasion: a meta-analysis. New Phytol. 2008;177:706–14.
Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–31.
Lipson DA, Raab TK, Parker M, Kelley ST, Brislawn CJ, Jansson J. Changes in microbial communities along redox gradients in polygonized Arctic wet tundra soils. Environ Microbiol Rep. 2015;7:649–57.
de Vries FT, Manning P, Tallowin JRB, Mortimer SR, Pilgrim ES, Harrison KA, et al. Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol Lett. 2012;15:1230–9.
Bartlett RJ, Ross DS. Colorimetric determination of oxidizable carbon in acid soil solutions. Soil Sci Soc Am J. 1988;1192:1191–2.
Brookes PC, Kragt JF, Powlson DS, Jenkinson DS. Chloroform fumigation and the release of soil nitrogen: the effects of fumigation time and temperature. Soil Biol Biochem. 1985;17:831–5.
Lipson DA, Schmidt SK, Monson RK. Links between microbial population dynamics and nitrogen availability in an alpine ecosistem. Ecology. 1999;80:1623–31.
Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–4.
Meyer F, Paarmann D, D’Souza M, Olson R, Glass E, Kubal M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008;9:386.
R Core Team. R: a language and environment for statistica computing. Vienna, Austria: R Foundation for Statistical Computing; 2017.
Chao Y, Yang Y, Ju F, Zhang X, Wu W, Zhang T. Metagenomic analysis reveals significant changes of microbial. Sci Rep. 2013;3:1–9.
Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46.
Zhang N, Liu W, Yang H, Yu X, Gutknecht JLM, Zhang Z, et al. Soil microbial responses to warming and increased precipitation and their implications for ecosystem C cycling. Oecologia. 2013;173:1125–42.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, Hara RBO et al. (2017). Vegan: community ecology package.
Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–4.
Zhao C, Miao Y, Yu C, Zhu L, Wang F, Jiang L, et al. Soil microbial community composition and respiration along an experimental precipitation gradient in a semiarid steppe. Sci Rep. 2016;6:24317.
Huxman TE, Snyder KA, Tissue D, Leffler AJ, Ogle K, Pockman WT, et al. Precipitation pulses and carbon fluxes in semiarid and arid ecosystems. Oecologia. 2004;141:254–68.
Potts M. Desiccation tolerance of prokaryotes. Microbiol Rev. 1994;58:755–805.
Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW. Soil microbial community responses to multiple experimental climate change drivers. Appl Environ Microbiol. 2010;76:999–1007.
Cruz-Martínez K, Rosling A, Zhang Y, Song M, Andersen GL, Banfield JF. Effect of rainfall-induced soil geochemistry dynamics on grassland soil microbial communities. Appl Environ Microbiol. 2012;78:7587–95.
Neilson JW, Califf K, Cardona C, Copeland A, Van Treuren W, Josephson KL, et al. Significant impacts of increasing aridity on the arid soil microbiome. mSystems. 2017;2:e00195–1. https://doi.org/10.1128/mSystems.00195-16.
Fierer N, Leff JW, Adams BJ, Nielsen UN, Thomas S, Lauber CL, et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. PNAS. 2012;109:21390–5.
Barnard RL, Osborne CA, Firestone MK. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013;7:2229–41.
Kidron GJ, Gutschick VP. Soil moisture correlates with shrub-grass association in the chihuahuan desert. Catena. 2013;107:71–79.
Pockman WT, Small EE. The influence of spatial patterns of soil moisture on the grass and shrub responses to a summer rainstorm in a Chihuahuan desert ecotone. Ecosystems. 2010;13:511–25.
Wang S, Fu BJ, Gao GY, Yao XL, Zhou J. Soil moisture and evapotranspiration of different land cover types in the Loess Plateau, China. Hydrol Earth Syst Sci. 2012;16:2883–92.
Kakumanu ML, Cantrell CL, Williams Ma. Microbial community response to varying magnitudes of desiccation in soil: a test of the osmolyte accumulation hypothesis. Soil Biol Biochem. 2013;57:644–53.
Shen B, Hohmann S, Jensen RG, Bohnert aH. Roles of sugar alcohols in osmotic stress adaptation. Replacement of glycerol by mannitol and sorbitol in yeast. Plant Physiol. 1999;121:45–52.
Karhu K, Auffret MD, Dungait JA, Hopkins DW, Prosser JI, Singh BK, et al. Temperature sensitivity of soil respiration rates enhanced by microbial community response. Nature. 2014;513:81–84.
Esch EH, Lipson DA, Cleland EE. Direct and indirect effects of shifting rainfall on soil microbial respiration and enzyme activity in a semi-arid system. Plant Soil. 2017;411:1–14.
Bradley BA, Houghton RA, Mustard JF, Hamburg SP. Invasive grass reduces aboveground carbon stocks in shrublands of the Western US. Glob Change Biol. 2006;12:1815–22.
Kramer TD, Warren RJ, Tang Y, Bradford Ma. Grass invasions across a regional gradient are associated with declines in belowground carbon pools. Ecosystems. 2012;15:1271–82.
Austin AT, Vivanco L. Plant litter decomposition in a semi-arid ecosystem controlled by photodegradation. Nature. 2006;442:555–8.
de Graaff M-A, Throop HL, Verburg PSJ, Arnone JA, Campos X. A synthesis of climate and vegetation cover effects on biogeochemical cycling in shrub-dominated drylands. Ecosystems. 2014;17:931–45.
Meisner A, De Deyn GB, de Boer W, van der Putten WH. Soil biotic legacy effects of extreme weather events influence plant invasiveness. Proc Natl Acad Sci. 2013;110:9835–8.
Schrama M, Bardgett RD. Grassland invasibility varies with drought effects on soil functioning. J Ecol. 2016;104:1250–8.
Thakur MP, Reich PB, Eddy WC, Stefanski A, Rich R, Hobbie SE, et al. Some plants like it warmer: increased growth of three selected invasive plant species in soils with a history of experimental warming. Pedobiologia. 2014;57:57–60.
Conlisk E, Swab R, Martínez-Berdeja A, Daugherty MP. Post-fire recovery in coastal sage scrub: seed rain and community trajectory. PLoS One. 2016;11:1–14.
DeSimone SA, Zedler PH. Shrub seedling recruitment in unburned California coastal sage scrub and adjacent grasslands. Ecology. 1999;80:2018–32.
Borken W, Matzner E. Reappraisal of drying and wetting effects on C and N mineralization and fluxes in soils. Glob Change Biol. 2009;15:808–24.
Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007. https://doi.org/10.1890/05-1839.
Capek P, Hříbalová V. Water-soluble polysaccharides from Salvia officinalis L. possessing immunomodulatory activity. Phytochemistry. 2004;65:1983–92.
Dennis PG, Miller AJ, Hirsch PR. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol. 2010;72:313–27.
Minic Z, Jouanin L. Plant glycoside hydrolases involved in??cell wall polysaccharide degradation. Plant Physiol Biochem. 2006;44:435–49.
Martinez-Garcia M, Brazel DM, Swan BK, Arnosti C, Chain PSG, Reitenga KG, et al. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of verrucomicrobia. PLoS One. 2012;7:1–11.
Carey CJ, Michael Beman J, Eviner VT, Malmstrom CM, Hart SC. Soil microbial community structure is unaltered by plant invasion, vegetation clipping, and nitrogen fertilization in experimental semi-arid grasslands. Front Microbiol. 2015. https://doi.org/10.3389/fmicb.2015.00466.
NOAA, NCEI. (2017). State of the climate: drought for Annual 2016.
Acknowledgements
We thank Melanie Merkley, Brenda García, Saad Al-Jaber, and Timothy J. Rowell for sample collection and laboratory assistance, Ellen H. Esch for statistical analyses, the Cleland Lab and Pablo Bryant for field experiment maintenance and Valerie Eviner for her intellectual contributions. This material is based upon work supported by the National Science Foundation Division of Environmental Biology grant No. DEB 1154082.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Pérez Castro, S., Cleland, E.E., Wagner, R. et al. Soil microbial responses to drought and exotic plants shift carbon metabolism. ISME J 13, 1776–1787 (2019). https://doi.org/10.1038/s41396-019-0389-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-019-0389-9
This article is cited by
-
Drought and vegetation restoration lead to shifts in soil microbial diversity and co-occurrence networks in California coastal prairie
Plant and Soil (2024)
-
High intensity perturbations induce an abrupt shift in soil microbial state
The ISME Journal (2023)
-
Extreme summers impact cropland and grassland soil microbiomes
The ISME Journal (2023)
-
Soil radiocarbon abundance in global forest ecosystems controlled by climate and soil properties
Plant and Soil (2023)
-
Asynchronous responses of microbial CAZymes genes and the net CO2 exchange in alpine peatland following 5 years of continuous extreme drought events
ISME Communications (2022)