Abstract
Persisters are transiently antibiotic-tolerant cells that complicate the treatment of bacterial infections. Both theory and experiments have suggested that persisters facilitate genetic resistance by constituting an evolutionary reservoir of viable cells. Here, we provide evidence for a strong positive correlation between persistence and the likelihood to become genetically resistant in natural and lab strains of E. coli. This correlation can be partly attributed to the increased availability of viable cells associated with persistence. However, our data additionally show that persistence is pleiotropically linked with mutation rates. Our theoretical model further demonstrates that increased survival and mutation rates jointly affect the likelihood of evolving clinical resistance. Overall, these results suggest that the battle against antibiotic resistance will benefit from incorporating anti-persister therapies.
Similar content being viewed by others
Introduction
Ever since the introduction of antibiotics in clinical practice, clinicians have been faced with the emergence and spread of genetic mutants resistant to the drugs’ therapeutic effects [1]. Resistance mechanisms allow bacteria to grow in elevated drug concentrations and are the main culprit for antibiotic treatment failure [2]. However, the awareness grows that other mechanisms besides genetic resistance can enable bacteria to survive in the presence of antibiotics. In addition to population-wide tolerance mechanisms [3] and shielding effects of in vivo physical niches [4], the formation of a small number of drug-tolerant cells or “persisters” in antibiotic-sensitive populations is being recognized as a frequent cause of treatment failure [5,6,7,8,9,10]. Theory and experiments have shown that depending on the treatment schedule, either resistance or tolerance/persistence can be the optimal bacterial strategy to cope with antibiotic stress. Resistance is likely to evolve when antibiotics are applied continuously and/or at low doses, while intermittent treatment with high drug concentrations tends to select for increased tolerance and persistence [11,12,13,14,15].
Importantly, tolerance and persistence have also been suggested to have the potential to act as a stepping stone towards the evolution of genetic resistance, as persisters provide a viable cell reservoir from which resistant mutants can emerge by horizontal gene transfer or de novo chromosomal mutations [16, 17]. This idea was first put forward more than 30 years ago, based on the frequent co-occurrence of penicillin tolerance and multiple drug resistance in clinical isolates [15, 18]. Later, antibiotic tolerance was indeed shown to be advantageous for the acquisition of resistance genes by transformation in Streptococcus pneumoniae [19]. Recent experiments have lent support to these early findings. Sebastian et al. showed that de novo resistant mutants arise with high frequency from the persister population in Mycobacterium tuberculosis [20], and the Balaban lab reported that tolerance, provoked by an extended lag time, increases the opportunity for resistance-conferring mutations to occur and spread in Escherichia coli populations under intermittent ampicillin exposure [21]. Furthermore, levels of persistence and resistance were shown to positively correlate across Pseudomonas strains [22].
However, the following two lines of reasoning suggest that the link between persistence and resistance is not merely based on the increased number of cells available for mutation, but that persistence could also be directly linked with altered mutation rates and increased probabilities of mutating into an antibiotic-resistant genotype. First, both mutation and persistence have been suggested to be induced by the activity of different stress responses [23,24,25]. Hence, high persister fractions and high mutation rates could coincide in stressful environments due to a partial overlap in underlying mechanisms [17]. Second, both high-persistence and mutator phenotypes are known to thrive in fluctuating environments and upon sudden environmental changes [11, 26,27,28,29,30,31,32], pointing to a possible evolutionary link between both traits. As yet, however, it is unknown whether persistence is associated with increased mutation rates and how this could affect the likelihood for genetic resistance to evolve.
Our aim was to investigate this outstanding issue. Therefore, we tested if persister fractions are correlated with the likelihood to form resistant colonies and if the emergence of resistant mutants from growing and non-growing populations is pleiotropically linked with persistence. We found that persistence is strongly linked with resistance development, even when the number of surviving cells is taken into account, revealing a pleiotropic link of persistence with the probability to acquire resistance-conferring mutations. Finally, we also developed a mathematical model to investigate how persistence and altered mutation rates would affect the emergence of antibiotic resistance in the setting of a typical clinical infection, where the immune system is also involved.
Materials and methods
Strains, culture conditions, and antibiotics
E. coli SX43, a derivative of BW25993, is the ancestor of oppB*, a high-persistence mutant resulting from a recent evolution experiment [11]. The de-evolved oppB* strain results from experimental evolution of the oppB* mutant in antibiotic-free conditions [11]. This strain has five additional mutations, causing decreased persister fractions as compared to the wild type strain. The hipA7 strain and its isogenic wild type used in this work were constructed by Pearl et al. [33] Strains were grown in Mueller-Hinton broth (shaking, 200 rpm) and on Mueller-Hinton or Luria-Bertani (LB) agar at 37 °C. For time-kill curves and persistence assays, final concentrations of 5 µg/ml ciprofloxacin, 5 µg/ml norfloxacin, and 400 µg/ml kanamycin were applied. Mannitol was used at a final concentration of 10 mM.
Minimum inhibitory concentrations (MICs), time-kill curves, and persistence assays
MICs were determined using the microdilution method. For time-kill curves of stationary phase populations, an overnight culture was diluted 1:100 in 100 ml fresh medium. After 16 h, the initial cell number was determined by plating. 1 ml aliquots were incubated with 5 µg/ml ciprofloxacin and the number of surviving cells was determined by plating at specified time points. Persistence assays were performed by determining survival after 5 h of antibiotic exposure. Mannitol was added 1 h before initiating antibiotic treatment.
Resistance evolution on agar plates
Overnight cultures were resuspended in 10 mM MgSO4 and 100 µl was plated on Mueller-Hinton agar containing antibiotics corresponding to 2x MIC (ciprofloxacin, norfloxacin, spectinomycin), 3x MIC (chloramphenicol), or 4x MIC (kanamycin). The number of initially plated cells was determined by plating on non-selective LB agar. Stable genetic resistance of the colonies was verified by streaking on fresh antibiotic-containing plates and MIC-testing (3–5 colonies per strain). All colonies grew after transfer and had increased MIC values, ranging from 2x to >64x the original MIC. Stability of ciprofloxacin in agar plates was tracked daily during long-term 37 °C incubation by streaking resistant (E. aerogenes ATCC 13048; MIC = 0.064 µg/ml) and non-resistant strains (SX43 and MG21) on incubated plates containing an initial concentration of 2x MIC. Even after 20 days of incubation, non-resistant strains were not able to grow on the plates, indicating that the effective concentration remained above the MIC. Time-kill kinetics of the plated population were determined by excising pieces of agar without resistant colonies, vigorously vortexing in 10 mM MgSO4, and plating on LB agar.
Resistance evolution in liquid medium
An overnight culture was diluted in fresh medium containing ciprofloxacin (2x MIC) to an inoculum size of 106 bacteria and incubated (37 °C, linear shaking) in an automated plate reader. Optical density (OD600) was measured every 15 min for 8 days. When a cell acquired a resistance-conferring mutation, its outgrowth could be monitored and the time until the mutational event was estimated from the resulting growth curve. MICs of colonies isolated from growth-positive populations at the end of the experiment were 2 to 16-fold increased, confirming genetic resistance. For each out of two independent experiments, 16 or 32 parallel populations were started from an overnight culture in individual wells, totaling 32 or 64 replicates for each strain.
Sequencing
Resistance-conferring mutations in gyrA were identified by PCR amplification and Sanger sequencing of the gyrA QRDR using primers described elsewhere [34].
Rifampicin-based fluctuation assays
From an overnight culture in Mueller-Hinton broth, 10 or 20 parallel cultures were grown from a small inoculum (5000 CFU/ml) in 96-well plates. After 24 h, the whole population was plated on LB agar supplemented with rifampicin (100 µg/ml). Additionally, 4 or 8 cultures per experiment were grown under the same conditions and used to estimate the final total cell number by plating on non-selective LB agar. Colonies on non-selective plates and rifampicin plates were scored after 24 and 48 hours of incubation, respectively. Experiments were repeated 2–4 times, yielding 30 or 40 replicate populations per strain per condition (SX43, n = 30; oppB*, n = 40; de-evolved oppB*, n = 30; MG21, n = 30; hipA7, n = 30).
Experiments with ECOR strains
20 ECOR strains were randomly selected from the total collection comprising 72 strains. For time-kill curves, strains were inoculated in 150 µl Mueller-Hinton broth in a 96-well plate and grown for 24 h. This culture was diluted 1:100 in 1.5 ml fresh medium in a 24-well plate and grown for 16 h. Cultures were subsequently diluted 1:100 in 1 ml fresh medium in 24-well plates and incubated for 2.5 h to obtain exponential growth. An aliquot was plated to determine the initial cell number, antibiotics were added, and the surviving cell number was determined at specified time points by spot plating. Exponential cultures were used because stationary phase killing was very limited and showed little variation between strains. Resistance evolution of ECOR isolates on ciprofloxacin agar was performed as stated above, except that 4x MIC instead of 2x MIC was used.
Mathematical model
To simulate bacterial population dynamics during antibiotic treatment of an infection, we constructed a mathematical model formulated as a system of differential equations containing the bacterial subpopulations depicted in Fig. 4a as well as a population of innate immune effector cells. This system was solved numerically in Wolfram Mathematica 10.0 using NDSolve[]. For different combinations of the switching rate and the mutation rate, the time until resistant cells take over the population (>108 cells) was calculated using NMinimize[] (default line search method). We chose realistic parameter values within the range reported for E. coli [11, 35] and in accordance with our own observations, rather than specific values for any antibiotic-strain combination. We verified that qualitatively, the model predictions did not change when parameter values were varied within a reasonable range (Fig. S12). A full description of the model and justification of the chosen parameter values is given in Supplementary Methods.
Data analysis and statistics
Time-kill curves
Biphasic killing parameters were determined by fitting a non-linear mixed model to the Log10-transformed, normally distributed number of surviving cells per milliliter (CFU/ml) using the R package nlme (https://cran.r-project.org/web/packages/nlme/index.html). The model was based on the equation Log10(CFU/ml) = Log10((N0−P0).e−kn.τ + P0.e−kp.τ), with τ the treatment time (in hours), N0 and P0 the number of normal and persister cells per ml at τ = 0, and kn and kp the rate at which normal and persister cells are killed (per hour) [36]. Strain was coded as a fixed factor and replicate was coded as a random factor. Best-fit parameters were compared statistically with Wald F-tests. For all time-kill curves, biphasic fits were superior over monophasic fits (Akaike Information Criterion).
Phylogeny of ECOR strains
A maximum likelihood phylogenetic tree of the 20 ECOR strains was inferred from three concatenated MLST markers (tonB, dinG, and DPP) identified with Phylomark [37] and aligned with ClustalW in Bio-Edit [38]. The tree was constructed using MEGA version 4 [39] and was based on the Kimura two-parameter model, using complete deletion of sites with gaps. Escherichia fergusonii was used as an outgroup to root the resulting tree.
Persistence and resistance in ECOR strains
Estimated persister fractions from biphasic fittings were used as a quantitative parameter for persistence. The average number of ciprofloxacin-resistant mutants per plated cell per day, from day 3 onwards, was used as a parameter reflecting resistance evolution. Both parameters were Log10-transformed in order to obtain normally distributed data. Based on the obtained phylogenetic tree, a phylogenetic generalized least squares (PGLS) regression assuming a Brownian evolution model was performed using the R packages ape (https://cran.r-project.org/web/packages/ape/index.html) and nlme (https://cran.r-project.org/web/packages/nlme/index.html). Outliers with a Cook’s distance greater than 1 were omitted from the analysis.
Resistance evolution on agar plates
A linear model was fitted to the number of new antibiotic-resistant colonies per plated cell (Log10-transformed to obtain normally distributed data) as a function of time. Significant differences between the model parameters were assessed with unpaired, two-sided t-tests. S.e.m.’s were calculated on the Log10-transformed data and transformed back to the original scale for representation in Fig. 2b and Fig. S4.
Resistance evolution in liquid medium
A cubic smoothing spline function was fitted to the OD600-values. The smoothing parameter was chosen by cross-validation. The time needed for a resistant mutant to emerge was estimated empirically as the time at which the first derivative of the fitted growth curve exceeded 0.1. A non-parametric Cox proportional hazards model was fitted to the right-censored distribution of lag times for each strain (n = 64 for SX43, oppB*, and de-evolved oppB*; n = 32 for MG21 and hipA7). Mutant and wild type lag times were compared by Wald tests on coefficients of the fitted model.
Determination of mutation rates
Data from rifampicin-based fluctuation assays and day 2 counts of ciprofloxacin plate assays were analyzed by fluctuation analysis with the R package ‘flan’ version 0.4, using a maximum likelihood function that takes into account final cell counts [40,41,42]. For experiments with ciprofloxacin, only 100 µl of a 5 ml culture was plated. To avoid underestimating the number of mutations in the total population, we used the correction factor proposed by Stewart [43]. Mutations in persister cells surviving exposure to lethal antibiotic stress were calculated as the number of resistant colonies divided by the viable cell count two days prior (mutations per cell per day), and the mutation rate was averaged over day 3-10. Viable cell counts were estimated from the fitted time-kill curves (Fig. S8). Statistical comparison was done with unpaired, one-sided t-tests with Welch’s correction in the case of unequal variances (F-test). Confidence intervals for relative differences between mutants and wild type were determined according to Fieller’s theorem.
Results
Persistence and the evolution of resistance are positively correlated in natural isolates and lab strains of E. coli
As a first probe for the link between persistence and the likelihood to evolve genetic resistance, we focused on a set of 20 natural isolates randomly picked from the ECOR collection, the standard panel of environmental diversity in E. coli [44]. The ECOR collection is ideal for this, as it displays substantial differences in persister fractions, whereas MIC variation is limited [45]. First, MICs were measured to confirm that all isolates are sensitive towards ciprofloxacin (all MICs ≤ 0.016 µg/ml, Table S1). Next, we determined two quantitative parameters: one reflecting persistence and one reflecting the likelihood for resistance to evolve. Persister fractions were derived from a biphasic exponential fit to time-kill data of cultures challenged with a high dose of ciprofloxacin (Fig. S1a, Table S2). Persistence is highly variable among strains (p < 0.0001, likelihood ratio test) and shows no correlation with MICs (Pearson’s r = 0.148, p = 0.535), corroborating previous findings [45]. To determine the rate of resistance evolution, we monitored the emergence of resistant colonies on ciprofloxacin-supplemented Mueller-Hinton agar (4x MIC) over a course of 20 days (Fig. S1b). Resistant mutants that were formed in the preculture (pre-exposure) were excluded by ignoring colonies that appeared during the first two days of incubation, as described previously [46,47,48]. That way, we focused only on mutants that arose de novo after antibiotic exposure, and excluded variation caused by the presence of mutants that originated during growth in the preculture. The average number of mutants per plated cell per day was subsequently used as a quantitative parameter for the evolution of resistance. In order to investigate the relationship between persistence and resistance evolution while accounting for phylogenetic non-independence among strains, we performed a phylogenetic generalized least squares (PGLS) regression (Fig. 1). Calculation of Cook’s distances indicated that four observations deviate strongly from the other data and severely influence the regression analysis (Fig. S2). The coefficient of determination (R²) for the 16 remaining isolates is 0.498, meaning that the variation in persister fractions explains 49.8 % of the variation in resistance evolution (p = 0.002). This result indicates that the persister fraction of a strain is a factor that strongly determines its tendency to evolve resistance.
Across natural E. coli isolates, persister fractions are strongly predictive of the likelihood to evolve genetic antibiotic resistance. Persister fractions (n = 3 per strain) and the average number of resistant mutants per plated cell per day (n = 5 per strain) on ciprofloxacin-containing Mueller-Hinton agar (4x MIC) are plotted for 16 ECOR isolates on a log-log scale. To account for the phylogenetic non-independence among strains, PGLS regression was used to fit a linear model onto the log-log transformed data (R² = 0.498, p = 0.002)
Next, we investigated if the same trend could be observed in well-characterized persistence mutants in E. coli lab strains. We selected three previously characterized persistence mutants: the hipA7 and oppB* high-persistence mutants, and the de-evolved oppB* mutant which shows a decreased persister fraction due to evolution in prolonged drug-free conditions. oppB* carries a single amino acid substitution in OppB, one of five subunits of the Opp oligopeptide permease system, which confers high persistence through a currently unknown mechanism [11]. In addition to the mutation in OppB, the de-evolved oppB* strain has five mutations, but it is currently unknown how these are responsible for the decreased persister fraction. hipA7 was the first recognized high-persistence allele and encodes the HipA toxin harboring two amino acid substitutions, causing persistence by inactivation of glutamyl-tRNA synthetase and subsequent induction of the stringent response [49, 50]. All strains are equally sensitive towards ciprofloxacin (MIC = 0.008 µg/ml, Table S4) and stationary phase time-kill curves of these mutants and their isogenic wild types show the expected persistence phenotypes (Fig. 2a, Table S3). As a first step, we wanted to thoroughly characterize resistance evolution in these strains.
Ciprofloxacin resistance development is increased in high-persistence mutants and decreased in a low-persistence mutant. a Ciprofloxacin time-kill curves of stationary phase cultures show increased persister fractions in hipA7 and oppB*, and decreased persistence in de-evolved oppB*. Individual data points from three independent replicates are shown. Lines represent a biphasic exponential fit to the data. Parameter estimates for persister fractions were compared with Wald F-tests. b Cumulative number of resistant mutants per plated cell on Mueller-Hinton agar supplemented with ciprofloxacin (2x MIC). Data points represent averages ± s.e.m. (n = 10). Statistical comparison was done by two-sided t-tests on coefficients of a log-linear model. c Cumulative proportion of wells with ciprofloxacin-containing Mueller-Hinton broth (2x MIC) in which resistant mutants emerged (n = 32-64). Statistical comparison was done by Wald tests on coefficients of a Cox proportional hazards model, to take into account censoring. d Amino acid changes caused by gyrA mutations detected in ciprofloxacin-resistant colonies. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001, and ns non-significant
As for the natural E. coli isolates, we quantified resistance evolution by following the emergence of resistant colonies on ciprofloxacin-containing Mueller-Hinton agar (2x MIC) over time. In agreement with our hypothesis, we found that the number of resistant colonies per plated cell is increased in hipA7 and oppB*, while it is decreased in de-evolved oppB* (Fig. 2b). We then investigated whether resistance evolution in high-persistence mutants is still increased when the number of persisters is decreased through the addition of mannitol, which is known to sensitize persisters to antibiotics [51]. Mannitol indeed suppresses the high-persistence phenotype of oppB*, while hipA7 still forms more persisters than its wild type under this condition (Fig. S3). Correspondingly, resistance evolution on mannitol-supplemented agar reverts to wild type levels in the case of oppB* but not hipA7 (Fig. S4).
In Gram-negative bacteria, the DNA gyrase subunit GyrA constitutes the main target for ciprofloxacin and other quinolones [52]. Mutations in a particular N-terminal region, the “quinolone resistance-determining region” (QRDR), are the most common mechanism of resistance, both in clinical isolates and in vitro evolution experiments [52,53,54]. To determine if mutations in GyrA are also involved in the evolution of antibiotic resistance in our experiments, we sequenced the gyrA QRDR in 10–11 ciprofloxacin-resistant colonies per strain and identified mutations causing amino acid changes at residues 81, 82, 83, and 87 in 35 out of 64 colonies (Fig. 2d). The remaining colonies bear no mutations in the gyrA QRDR, but presumably have mutations in another gyrA region or in parC, encoding the topoisomerase IV subunit A [52]. The amino acid substitutions that we found are commonly observed in clinical isolates of quinolone-resistant E. coli as well as other Enterobacteriaceae and Gram-positive organisms [52, 55, 56].
To further confirm our finding of persistence being strongly linked to the likelihood of evolving antibiotic resistance, we also quantified antibiotic resistance development using an independent approach. We inoculated a population of 106 bacteria in ciprofloxacin-containing Mueller-Hinton broth (2x MIC) and measured the time until the emergence of resistance by tracking growth. As expected, the number of populations that had evolved resistance after eight days was strongly linked with the persistence phenotypes: 29/32 for hipA7 (14/32 for the wild type), 48/64 for oppB* and 15/64 for de-evolved oppB* (37/64 for the wild type) (Fig. 2c). Hence, the rate of resistance evolution is increased in hipA7 and oppB*, and decreased in de-evolved oppB*.
To assess the generality of our observations, we further tracked the emergence of resistant colonies on Mueller-Hinton agar supplemented with other antibiotics: norfloxacin, kanamycin, chloramphenicol, and spectinomycin. MICs of these antibiotics are given in Table S4. Norfloxacin and kanamycin are bactericidal drugs, and we confirmed the high-persistence phenotypes of hipA7 and oppB* upon treatment with these antibiotics (Fig. S5). The persister fraction of de-evolved oppB* is only very slightly decreased as compared to the wild type. Chloramphenicol and spectinomycin are bacteriostatic antibiotics, which inhibit E. coli growth but do not induce killing. The positive correlation between persister fractions and resistance development holds true for each antibiotic, except for spectinomycin. Only very few resistant colonies appeared on spectinomycin-supplemented plates, resulting in large variations and no significant differences for oppB* and de-evolved oppB* as compared to their wild type (Fig. S6).
A link between persistence and mutation rates
The observed correlation between persistence and resistance development could be due to an increased viable cell reservoir, a pleiotropic effect of persistence on mutation rates, or a combination of both. To investigate the contribution of mutagenesis, we compared mutation rates in the well-defined high and low-persistence mutants. Resistant mutants can be generated from growing bacteria as well as from non-growing but surviving bacteria exposed to growth-restricted conditions [47]. Based on earlier studies, resistant colonies that emerge on antibiotic-containing agar during the first two days of incubation can be traced back to resistant mutants that emerged during growth in the preculture [46, 47]. Thus, from the number of colonies on day 2, the mutation rate during growth (mutations per cell per division) was estimated by fluctuation analysis. The results from these experiments demonstrate that the mutation rate is increased in hipA7 (2.8-fold) and oppB* (5.0-fold), while it is decreased in de-evolved oppB* (2.0-fold) (Fig. 3a, Table S5). We further confirmed these results using a rifampicin-based fluctuation assay to determine the mutation rate at another, independent genomic locus (Fig. S7, Table S5).
Persistence is correlated with elevated mutation rates. a Mutation rate (mutations per cell per division) of growing cells determined from the number of ciprofloxacin-resistant mutants after two days using fluctuation analysis, relative to the wild type. Error bars show 95% c.i. (n = 10), p-values are from one-sided t-tests. b Mutations in non-growing cells surviving lethal antibiotic exposure (mutations per cell per day), relative to the wild type. Error bars show 95% c.i.’s (n = 10), p-values are from one-sided t-tests. *p ≤ 0.1; **p ≤ 0.05; ***p ≤ 0.01
Apart from the resistant colonies that appear within two days of incubation and acquired a mutation during growth in the preculture, mutants are also formed de novo from the surviving subpopulation on the ciprofloxacin-containing agar plate. To quantify the mutation rate of cells from the surviving reservoir of non-growing persisters, the number of resistant colonies has to be normalized for the remaining number of viable cells on the plate. To estimate the size of the viable cell reservoir, we measured time-kill kinetics of the plated populations by determining the number of surviving cells at different time points after plating (Fig. S8). In agreement with earlier reports [46,47,48], a biphasic killing pattern was observed for all strains except hipA7. The bulk of the population (>99.9%) is killed within the first day after plating, while a small subpopulation of cells is killed at a much slower rate. When we correct the number of resistant colonies for the number of surviving cells in the persister reservoir, we still observe a correlation between persistence and resistance. The average mutation rate per viable cell per day is significantly increased in hipA7 and oppB*, and significantly lower in de-evolved oppB*, compared to their respective wild types (Fig. 3b). Together, these experiments reveal that persistence correlates with mutation rates under various growth conditions and in this way has a strong effect on the likelihood to evolve genetic resistance.
Increased survival and mutation rates jointly drive evolution of clinical resistance
To assess the implications of our findings under clinical conditions, we developed a mathematical model to simulate antibiotic treatment of an in vivo bacterial infection. In this model, bacteria continuously switch between the normal and persister states following switching rates a and b. Growth and killing of non-persisters is related to the antibiotic concentration according to a pharmacodynamic Hill function [35]. Persisters are assumed to be non-dividing and recalcitrant to killing by antibiotics. Both normal and persister cells can be killed by the action of an innate immune response, although the killing rate is taken to be lower for persister cells [57, 58]. As the immune system alone is not sufficient to control this infection within a reasonable amount of time, bacterial clearance is highly dependent on the action of the antibiotic [59]. The antibiotic is added every 12 h at a peak concentration of 25x MIC (Amax) and the effective concentration declines exponentially with a half-life of 2.5 h (Fig. S9). As such, the model incorporates realistic clinical pharmacokinetics and pharmacodynamics for a broad range of antibiotics (see Supplementary Methods and Table S6 for details).
Our model focuses on resistance evolution via chromosomal mutations. In this case, clinical resistance is mostly achieved by multiple mutations accumulated in a stepwise fashion [60]. Therefore, we implemented resistance evolution as a gradual process, where subsequent mutations generate mutants showing 4, 20, and 50-fold increased MIC values. Replicating cells develop resistance according to the mutation rate µgrowing, which can be increased up to twofold by the presence of sublethal concentrations of antibiotics. Non-dividing persisters acquire resistance-conferring mutations at a constant rate µpersisters. A schematic diagram showing the possible interconversions between all different phenotypes and genotypes is shown in Fig. 4a.
A mathematical simulation model shows that increased persistence and mutation rates jointly drive the evolution of resistance under clinical conditions. a Schematic diagram of the mathematical model showing all possible transitions between the different genotypes and phenotypic states. b Population dynamics for wild type and resistant cells during antibiotic treatment for increasing persister fractions with or without associated increased mutation rates. The separate dynamics of all different subpopulations under the same sets of parameters are given in Fig. S10. c Heat map of the time at which the resistant cells take over the population (>108 resistant cells), as a function of the mutation rate and the persister fraction (switching rate). The mutation rate is shown for normal cells, the mutation rate for persisters is 104-fold higher. See text and Supplementary Methods for details on the mathematical model
Fig. 4b depicts bacterial population dynamics for pathogens exhibiting different persister fractions, with and without an associated increase in mutation rates (for separate dynamics of all different subpopulations, see Fig. S10). In the baseline situation, the infection is cleared within 5 days of antibiotic treatment. A small subpopulation of resistant cells emerges, but it cannot acquire the multiple subsequent mutations necessary to obtain high-level resistance, and therefore it does not impair treatment success. When we increase switching to the persister state 10-fold, the infection is still cured, although bacterial clearance is delayed. Similarly, increasing the mutation rates by a factor in line with our experimental findings, i.e., twofold for µgrowing and fourfold for µpersisters, has no effect on the infection dynamics. However, when we concomitantly increase persistence and mutation rates, antibiotic therapy fails as highly resistant mutants can now take over the population. Only upon a higher, 50-fold increase in persister switching, high-level resistance can already emerge with baseline mutation rates, and even then, an associated increase in mutation rate causes high-level resistant mutants to take over the population sooner. These examples clearly illustrate that increased persister fractions and mutation rates jointly drive the evolution of genetic antibiotic resistance. To investigate this effect over a larger range of parameter values, we calculated the time it takes for the resistant population to reach a density of 108 cells, given different combinations of persister fractions and mutation rates. The resulting heat map confirms that both parameters simultaneously drive the evolution of resistance (Fig. 4c).
To verify that a mutant with a pleiotropically linked persister fraction and mutation rate is actually favored over the wild type, we calculated the invasion potential of rare mutants with different combinations of switching rates and mutation rates in a wild type population. The results of this simulation suggest that the selective advantage of a mutant is proportional to its persister fraction as well as its mutation rate. Moreover, mutants with high values for both parameters exhibit the strongest invasion potential, pointing towards a possible evolutionary link between persistence and mutation rates (Fig. S11).
Discussion
To date, many lines of evidence strongly suggest that persisters contribute to treatment failure of bacterial infections [6]. In this paper, we show that the clinical burden of persisters probably extends beyond their direct role in chronic and recurrent infections because they strongly contribute to the evolution of genetic resistance. By tracking the appearance of resistant colonies on ciprofloxacin-containing agar, we found that persistence is a crucial factor driving the acquisition of genetic resistance by chromosomal mutations. Targeted sequencing revealed that the responsible mutations are similar to those encountered in fluoroquinolone-resistant clinical isolates in a number of bacterial species [52, 55, 56]. The effect of persistence was also not only observed with ciprofloxacin, but is valid for various antibiotics with different modes of action. Moreover, the quantitative relationship between persister fractions and ciprofloxacin resistance evolution in 80% of the tested natural isolates of E. coli suggests that the link is intrinsic and widespread.
The obvious mechanism by which persistence can facilitate resistance development is by providing more viable cells eligible for mutation. The number of surviving cells will then influence the likelihood that resistance emerges. Indeed, Levin and Rozen illustrated mathematically that increasing the number of persisters augments the likelihood of evolving resistance [16]. Recent experimental work has confirmed that resistant mutants can be generated de novo from the persister population during antibiotic treatment of M. tuberculosis [20]. Levin-Reisman et al. showed that mutants exhibiting increased levels of ampicillin tolerance or persistence evolved resistance earlier than the wild type [21].
In addition to this straightforward mechanism, we hypothesized that persistence and resistance could also be linked in a less obvious way. Indeed, two different lines of reasoning suggest that persistence correlates with mutation rates. First, the involvement of stress responses in persistence points to a link with mutation rates of cells growing in stressful environments [17]. According to a recent study, this effect could be especially important when the stress response is activated by DNA damage, because erroneous repair of DNA damage is a major source of mutations [61]. A second argument stems from evolutionary theory. Mathematical models indicate that persistence may have evolved as a bet-hedging strategy in environments characterized by fluctuating episodes of stress [26, 62,63,64]. Recent experimental evidence lends strong support to these theoretic considerations, showing that persistence is disadvantageous in constant environments but favored during periodic stress [11, 13, 14]. Mathematical models have also been used to show that high mutation rates are favored when the environment shows periodic changes [27, 28]. However, models describing mutagenesis are mathematically equivalent to models of phenotypic switching, as they consider a single fitness-determining selective locus with only two possible allelic states that can mutate back and forth. Nevertheless, both theory and experiments have shown the adaptive benefit of increased mutation rates to cope with stress or a sudden environmental change. Maladapted genotypes are predicted to experience selection for increased mutation rates in both constant and fluctuating environments [32]. Evolution experiments within laboratory [29] or host environments [31] confirmed these theoretical predictions by showing selection for increased mutation rates in conditions of near-lethal stress and colonization of a new host. Moreover, it was shown that antibiotic treatment can select mutator alleles, especially during chronic infections that are repeatedly challenged with antibiotics [30].
We tested this putative association between persistence and mutation by directly measuring mutation rates in high- and low-persistence mutants. Using fluctuation assays with ciprofloxacin and rifampicin selection, we found that persistence is pleiotropically linked with mutation rates in growing cells. Moreover, the number of resistant colonies emerging from the viable cell reservoir shows the same correlation, even when a correction for the number of surviving cells is applied. Thus, persister cells formed by high-persistence strains have an increased likelihood to acquire resistance-conferring mutations, while persisters from low-persistence strains are less likely to mutate and hence to acquire resistance. The positive association between persistence and mutation rates is important, since even modest increases in the mutational supply can dramatically improve evolvability [65,66,67]. Interestingly, these findings also imply that the clinical relevance of persistence is not limited to infections treated with bactericidal antibiotics. High-persistence strains, which have repeatedly been identified in chronic infections [68, 69], can even complicate treatment in the absence of bactericidal drugs because they are more likely to evolve resistance due to their increased mutation rates.
The continuous emergence of resistant colonies on antibiotic-containing agar plates over the course of several days or weeks is usually explained by de novo mutation in non-growing, surviving cells [46,47,48]. Indeed, spontaneous or antibiotic-induced DNA damage in non-growing cells may lead to mutations because, even in the absence of chromosomal replication, these cells still display DNA turnover [70, 71]. On the other hand, others have proposed that these and similar observations do not involve mutations in the absence of growth, but can instead be explained by pre-existing small-effect mutants that grow slowly and give rise to faster-growing mutants that subsequently form colonies [72]. These two contrasting views are subject of considerable debate [71,72,73]. Cirz et al. have shown that deletion or mutation of several genes can dramatically decrease late colony formation on ciprofloxacin-containing agar plates, without affecting the number of early colonies, fitness under normal or growth-restricted conditions, or the selective advantage of resistance-conferring mutations [46]. These results indicate that de novo mutation in non-growing cells is the most likely explanation for the gradual accumulation of colonies on ciprofloxacin plates.
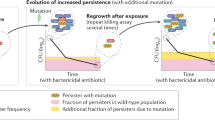
Taken together, our results lead us to propose a model in which two factors constitute the link between persistence and resistance. When a population is treated with bactericidal antibiotics (Fig. 5, red frame, non-growing conditions), only persisters will survive. Increased persistence leads to more cells in the viable cell reservoir and an increased likelihood for the occurrence of resistance-conferring mutations. Moreover, the mutation rate of persisters is increased in high-persistence strains. When antibiotics are absent or present at low concentrations (Fig. 5, green frame, growing conditions), the mutation rate of growing cells is correlated with persistence. Elevated mutation rates in turn increase the likelihood for the emergence of resistance.
Proposed model explaining how persistence accelerates the evolution of genetic resistance. Persistence refers to the fraction of cells in a sensitive population that survives otherwise lethal antibiotic exposure. High persistence accelerates the evolution of antibiotic resistance by increasing the number of viable cells available for mutation, thus increasing the likelihood for a resistance-conferring mutation to arise. Additionally, persistence is linked with higher mutation rates in growing cells and cells that survive lethal antibiotic exposure but cannot grow (persisters). Together, these two factors have the potential to accelerate the evolution of antibiotic resistance in high-persistence strains in different environments with or without antibiotics. Red depicts environments where the antibiotic concentration is above the MIC, while green depicts environments with subinhibitory or zero antibiotic concentrations
To explore the implications of our findings in a clinical context, we developed a mathematical model simulating bacterial infection dynamics during antibiotic treatment. Our model significantly extends on previous modeling efforts by incorporating step-wise resistance evolution and a flexible mutation rate under antibiotic stress, leading to a model that is closer to a realistic clinical situation. Under conditions that mimic those encountered in the clinic and using parameter values in range for E. coli and in line with our findings, the model predicts that the impact of increased persistence on the evolution of resistance is considerably greater when increased persister fractions are correlated with increased mutation rates. As our simulations also demonstrate, these mutants with pleiotropically linked high persistence and mutation rates are likely to emerge in fluctuating conditions.
Current efforts to tackle the problem of antibiotic resistance are insufficient, as antibiotic-resistant pathogens pose a growing public health problem [2, 74]. As the pipeline of new antibiotics active against these pathogens is very thin, inhibiting the evolution of resistance could be an important part of the solution. Indeed, directly intervening with evolution is regarded as a promising therapeutic strategy in bacterial infections and beyond [75,76,77]. Antibiotic resistance evolution is a complex process, influenced by many factors including stress responses, mismatch repair, iron homeostasis, and translation fidelity [25, 65]. Proposed strategies to inhibit antibiotic resistance evolution include combination therapy [78], drug cycling protocols [79], interference with the SOS response [46], and small-molecule RecA inhibitors [48]. Our results indicate that bacterial persistence also plays a pivotal role in the development of genetic antibiotic resistance and could represent a novel and innovative way to combat it. Interestingly, it was recently shown that the molecular mechanisms underlying drug-tolerant persisters in cancer could be a novel and promising target to prevent acquired therapy resistance [80, 81]. In conclusion, we propose that anti-persister strategies may be an important part of the solution to the ongoing antibiotic crisis, not only by shortening treatment durations but also by slowing down resistance development.
References
Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–33.
World Health Organization. Antimicrobial resistance: global report on surveillance. WHO press, Geneva, Switzerland; 2014.
Brauner A, Fridman O, Gefen O, Balaban NQ. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol. 2016;14:320–30.
Bumann D. Heterogeneous host-pathogen encounters: act locally, think globally. Cell Host Microbe. 2015;17:13–19.
Conlon BP. Staphylococcus aureus chronic and relapsing infections: evidence of a role for persister cells. Bioessays. 2014;36:991–6.
Michiels JE, Van den Bergh B, Verstraeten N, Michiels J. Molecular mechanisms and clinical implications of bacterial persistence. Drug Resist Updat. 2016;29:76–89.
Van den Bergh B, Michiels JE, Fauvart M, Michiels J. Should we develop screens for multidrug tolerance? Expert Rev Anti Infect Ther. 2016;14:613–6.
Van den Bergh B, Fauvart M, Michiels J. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol Rev. 2017;41:219–51.
Harms A, Maisonneuve E, Gerdes K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science. 2016;354:aaf4268.
Fisher RA, Gollan B, Helaine S. Persistent bacterial infections and persister cells. Nat Rev Microbiol. 2017;15:453–64.
Van den Bergh B, Michiels JE, Wenseleers T, Windels EM, Vanden Boer P, Kestemont D, et al. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat Microbiol. 2016;1:16020.
Michiels JE, Van den Bergh B, Verstraeten N, Fauvart M, Michiels J. In vitro emergence of high persistence upon periodic aminoglycoside challenge in the ESKAPE pathogens. Antimicrob Agents Chemother. 2016;60:4630–7.
Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature. 2014;513:418–21.
Mechler L, Herbig A, Paprotka K, Fraunholz M, Nieselt K, Bertram R. A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrob Agents Chemother. 2015;59:5366–76.
Moreillon P, Tomasz A. Penicillin resistance and defective lysis in clinical isolates of pneumococci: evidence for two kinds of antibiotic pressure operating in the clinical environment. J Infect Dis. 1988;157:1150–7.
Levin BR, Rozen DE. Non-inherited antibiotic resistance. Nat Rev Microbiol. 2006;4:556–62.
Cohen NR, Lobritz MA, Collins JJ. Microbial persistence and the road to drug resistance. Cell Host Microbe. 2013;13:632–42.
Liu HH, Tomasz A. Penicillin tolerance in multiply drug-resistant natural isolates of Streptococcus pneumoniae. J Infect Dis. 1985;152:365–72.
Novak R, Henriques B, Charpentier E, Normark S, Tuomanen E. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature. 1999;399:590–3.
Sebastian J, Swaminath S, Nair RR, Jakkala K, Pradhan A, Ajitkumar P. De novo emergence of genetically resistant mutants of Mycobacterium tuberculosis from the persistence phase cells formed against antituberculosis drugs in vitro. Antimicrob Agents Chemother. 2016;61:AAC.01343-16.
Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. Antibiotic tolerance facilitates the evolution of resistance. Science. 2017;355:826–30.
Vogwill T, Comfort AC, Furió V, MacLean RC. Persistence and resistance as complementary bacterial adaptations to antibiotics. J Evol Biol. 2016;29:1223–33.
Bjedov I, Tenaillon O, Gérard B, Souza V, Denamur E, Radman M, et al. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–9.
Al Mamun, AAM Lombardo M, Shee C, Lisewski AM, Gonzalez C, et al. Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science. 2012;338:1344–8.
Poole K. Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria. Trends Microbiol. 2012;20:227–34.
Kussell E, Kishony R, Balaban NQ, Leibler S. Bacterial persistence: a model of survival in changing environments. Genetics. 2005;169:1807–14.
Carja O, Liberman U, Feldman MW. Evolution in changing environments: Modifiers of mutation, recombination, and migration. Proc Natl Acad Sci. 2014;111:17935–40.
Ishii K, Matsuda H, Iwasa Y, Sasaki A. Evolutionarily stable mutation rate in a periodically changing environment. Genetics. 1989;121:163–74.
Swings T, Van den Bergh B, Wuyts S, Oeyen E, Voordeckers K, Verstrepen KJ, et al. Adaptive tuning of mutation rates allows fast response to lethal stress in Escherichia coli. eLife. 2017;6:1–24.
Giraud A, Matic I, Radman M, Fons M, Taddei F. Mutator bacteria as a risk factor in treatment of infectious diseases. Antimicrob Agents Chemother. 2002;46:863–5.
Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, et al. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science. 2001;291:2606–8.
Ram Y, Hadany L. The evolution of stress-induced hypermutation in asexual populations. Evolution. 2012;66:2315–28.
Pearl S, Gabay C, Kishony R, Oppenheim A, Balaban NQ. Nongenetic individuality in the host-phage interaction. PLoS Biol 2008;6:0957–64.
Wang H, Dzink-Fox J, Chen M, Levy SB. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother. 2001;45:1515–21.
Regoes RR, Wiuff C, Zappala RM, Garner KN, Baquero F, Levin BR. Pharmacodynamic functions: a multiparameter approach to the design of antibiotic treatment regimens. Antimicrob Agents Chemother. 2004;48:3670–6.
Stepanyan K, Wenseleers T, Duéñez-Guzmán EA, Muratori F, Van den Bergh B, Verstraeten N, et al. Fitness trade-offs explain low levels of persister cells in the opportunistic pathogen Pseudomonas aeruginosa. Mol Ecol. 2015;24:1572–83.
Sahl JW, Matalka MN, Rasko DA. Phylomark, a tool to identify conserved phylogenetic markers from whole-genome alignments. Appl Environ Microbiol. 2012;78:4884–92.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98.
Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9.
Hamon A, Ycart B. Statistics for the Luria-Delbrück distribution. Electron J Stat. 2012;6:1251–72.
Ycart B, Veziris N. Unbiased estimation of mutation rates under fluctuating final counts. PLoS ONE. 2014;9:e101434.
Mazoyer A, Drouilhet R, Despréaux S, Ycart B. flan: An R package for inference on mutation models. R J. 2017;9:334–51.
Stewart FM. Fluctuation analysis: the effect of plating efficiency. Genetica. 1991;84:51–55.
Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–3.
Stewart B, Rozen DE. Genetic variation for antibiotic persistence in Escherichia coli. Evolution. 2011;66:933–9.
Cirz RT, Chin JK, Andes DR, de Crécy-Lagard V, Craig WA, Romesberg FE. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:1024–33.
Riesenfeld C, Everett M, Piddock LJ, Hall BG. Adaptive mutations produce resistance to ciprofloxacin. Antimicrob Agents Chemother. 1997;41:2059–60.
Alam MK, Alhhazmi A, DeCoteau JF, Luo Y, Geyer CR. RecA inhibitors potentiate antibiotic activity and block evolution of antibiotic resistance. Cell Chem Biol. 2016;23:381–91.
Kaspy I, Rotem E, Weiss N, Ronin I, Balaban NQ, Glaser G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat Commun. 2013;4:3001.
Germain E, Castro-Roa D, Zenkin N, Gerdes K. Molecular mechanism of bacterial persistence by HipA. Mol Cell. 2013;52:248–54.
Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–20.
Piddock LJ. Mechanisms of fluoroquinolone resistance: an update 1994-1998. Drugs. 1999;58:11–18.
Oz T, Guvenek A, Yildiz S, Karaboga E, Tamer YT, Mumcuyan N, et al. Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. Mol Biol Evol. 2014;31:2387–401.
Lázár V, Nagy I, Spohn R, Csörgő B, Györkei Á, Nyerges Á, et al. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat Commun. 2014;5:4352.
Conrad S, Oethinger M, Kaifel K, Klotz G, Marre R, Kern WV. gyrA mutations in high-level fluoroquinolone-resistant clinical isolates of Escherichia coli. J Antimicrob Chemother. 1996;38:443–55.
Weigel LM, Steward CD, Tenover FC. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob Agents Chemother. 1998;42:2661–7.
Ankomah P, Levin BR. Exploring the collaboration between antibiotics and the immune response in the treatment of acute, self-limiting infections. Proc Natl Acad Sci USA. 2014;111:8331–8.
Putrinš M, Kogermann K, Lukk E, Lippus M, Varik V, Tenson T. Phenotypic heterogeneity enables uropathogenic Escherichia coli to evade killing by antibiotics and serum complement. Infect Immun. 2015;83:1056–67.
Levin BR, Baquero F, Ankomah P, McCall IC. Phagocytes, antibiotics, and self-limiting bacterial infections. Trends Microbiol. 2017;25:878–92.
Normark BH, Normark S. Evolution and spread of antibiotic resistance. J Intern Med. 2002;252:91–106.
Yaakov G, Lerner D, Bentele K, Steinberger J, Barkai N. Coupling phenotypic persistence to DNA damage increases genetic diversity in severe stress. Nat Ecol Evol. 2017;1:16.
Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–8.
Patra P, Klumpp S. Population dynamics of bacterial persistence. PLoS ONE. 2013;8:e62814.
Gardner A, West SA, Griffin AS. Is bacterial persistence a social trait? PLoS ONE. 2007;2:e752.
Méhi O, Bogos B, Csörgo B, Pál C. Genomewide screen for modulators of evolvability under toxic antibiotic exposure. Antimicrob Agents Chemother. 2013;57:3453–6.
Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–20.
Long H, Miller SF, Strauss C, Zhao C, Cheng L, Ye Z, et al. Antibiotic treatment enhances the genome-wide mutation rate of target cells. Proc Natl Acad Sci USA. 2016;113:E2498–E2505.
LaFleur MD, Qi Q, Lewis K. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob Agents Chemother. 2010;54:39–44.
Mulcahy LR, Burns JL, Lory S, Lewis K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol. 2010;192:6191–9.
Bridges BA. DNA turnover and mutation in resting cells. Bioessays. 1997;19:347–52.
Williams AB, Foster PL. Stress-induced mutagenesis. EcoSal Plus. 2012;5:1–29.
Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. Origin of mutations under selection: the adaptive mutation controversy. Annu Rev Microbiol. 2006;60:477–501.
Rosenberg SM, Shee C, Frisch RL, Hastings PJ. Stress-induced mutation via DNA breaks in Escherichia coli: A molecular mechanism with implications for evolution and medicine. Bioessays. 2012;34:885–92.
O’Neill J. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Review on Antimicrobial Resistance. 2014.
Rosenberg SM, Queitsch C. Combating evolution to fight disease. Science. 2014;343:1088–9.
Cirz RT, Romesberg FE. Controlling mutation: intervening in evolution as a therapeutic strategy. Crit Rev Biochem Mol Biol. 2007;42:341–54.
Goldberg DE, Siliciano RF, Jacobs WR. Outwitting evolution: fighting drug-resistant TB, malaria, and HIV. Cell. 2012;148:1271–83.
Fischbach MA. Combination therapies for combating antimicrobial resistance. Curr Opin Microbiol. 2011;14:519–23.
Imamovic L, Sommer MOA. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci Transl Med. 2013;5:204ra132.
Oxnard GR. The cellular origins of drug resistance in cancer. Nat Med. 2016;22:232–4.
Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247–50.
Acknowledgements
We thank Chris Michiels (KU Leuven) for providing ECOR strains, Abram Aertsen (KU Leuven) for sharing the hipA7 strain, and Bernard Ycart and Adrien Mazoyer (Grenoble Alpes University) for their assistance with fluctuation analysis. EMW and BVdB are Research Foundation—Flanders (FWO)-fellows and JEM is a fellow of the Agency for Innovation by Science and Technology (IWT). This research was funded by the KU Leuven Research Council (PF/10/010; C16/17/006), the Belgian Science Policy Office (IAP P7/28), FWO (G047112N; G0B2515N; G055517N), and the Flemish Institute for Biotechnology (VIB).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Windels, E.M., Michiels, J.E., Fauvart, M. et al. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J 13, 1239–1251 (2019). https://doi.org/10.1038/s41396-019-0344-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-019-0344-9
This article is cited by
-
A role for the stringent response in ciprofloxacin resistance in Pseudomonas aeruginosa
Scientific Reports (2024)
-
Macrophage internalization creates a multidrug-tolerant fungal persister reservoir and facilitates the emergence of drug resistance
Nature Communications (2023)
-
Intermittent antibiotic treatment of bacterial biofilms favors the rapid evolution of resistance
Communications Biology (2023)
-
Targeted Hypermutation as a Survival Strategy: A Theoretical Approach
Acta Biotheoretica (2023)
-
Coumarin: A Renowned Moiety for Gram-Negative Escherichia coli – a Review
Pharmaceutical Chemistry Journal (2023)