Abstract
Purpose
To develop nomograms that predict the detection of clinically significant prostate cancer (csPCa, defined as ≥GG2 [Grade Group 2]) at diagnostic biopsy based on multiparametric prostate MRI (mpMRI), serum biomarkers, and patient clinicodemographic features.
Materials and methods
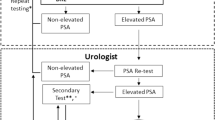
Nomograms were developed from a cohort of biopsy-naïve men presenting to our 11-hospital system with prostate specific antigen (PSA) of 2–20 ng/mL who underwent pre-biopsy mpMRI from March 2018-June 2021 (n = 1494). The outcomes were the presence of csPCa and high-grade prostate cancer (defined as ≥GG3 prostate cancer). Using significant variables on multivariable logistic regression, individual nomograms were developed for men with total PSA, % free PSA, or prostate health index (PHI) when available. The nomograms were both internally validated and evaluated in an independent cohort of 366 men presenting to our hospital system from July 2021-February 2022.
Results
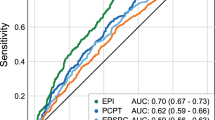
1031 of 1494 men (69%) underwent biopsy after initial evaluation with mpMRI, 493 (47.8%) of whom were found to have ≥GG2 PCa, and 271 (26.3%) were found to have ≥GG3 PCa. Age, race, highest PIRADS score, prostate health index when available, % free PSA when available, and PSA density were significant predictors of ≥GG2 and ≥GG3 PCa on multivariable analysis and were used for nomogram generation. Accuracy of nomograms in both the training cohort and independent cohort were high, with areas under the curves (AUC) of ≥0.885 in the training cohort and ≥0.896 in the independent validation cohort. In our independent validation cohort, our model for ≥GG2 prostate cancer with PHI saved 39.1% of biopsies (143/366) while only missing 0.8% of csPCa (1/124) with a biopsy threshold of 20% probability of csPCa.
Conclusions
Here we developed nomograms combining serum testing and mpMRI to help clinicians risk stratify patients with elevated PSA of 2–20 ng/mL who are being considered for biopsy. Our nomograms are available at https://rossnm1.shinyapps.io/MynMRIskCalculator/ to aid with biopsy decisions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Data may be available for review and secondary analysis upon request as per institutional guidelines and policies.
References
American Cancer Society. Cancer facts & figures 2022. Atlanta: American Cancer Society; 2022. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2022/2022-cancer-facts-and-figures.pdf.
Loeb S, Sanda MG, Broyles DL, Shin SS, Bangma CH, Wei JT, et al. The prostate health index selectively identifies clinically significant prostate cancer. J Urol. 2015;193:1163.
Padhani AR, Petralia G, Sanguedolce F. Magnetic resonance imaging before prostate biopsy: time to talk. Eur Urol. 2016;69:1–3.
Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389:815–22.
Kasivisvanathan V, Rannikko AS, Borghi M, Panebianco V, Mynderse LA, Vaarala MH, et al. MRI-targeted or standard biopsy for prostate-cancer diagnosis. N Engl J Med. 2018;378:1767–77.
Park KJ, Choi SH, Lee JS, Kim JK, Kim MH, Jeong IG. Risk stratification of prostate cancer according to PI-RADS® Version 2 categories: meta-analysis for prospective studies. J Urol. 2020;204:1141–9.
Oerther B, Engel H, Bamberg F, Sigle A, Gratzke C, Benndorf M. Cancer detection rates of the PI-RADSv2.1 assessment categories: systematic review and meta-analysis on lesion level and patient level. Prostate Cancer Prostatic Dis. 2021. https://doi.org/10.1038/s41391-021-00417-1.
Kinnaird A, Brisbane W, Kwan L, Priester A, Chuang R, Barsa DE, et al. A prostate cancer risk calculator: use of clinical and magnetic resonance imaging data to predict biopsy outcome in North American men. Can Urol Assoc J. 2022;16:E161.
Alberts AR, Roobol MJ, Verbeek JFM, Schoots IG, Chiu PK, Osses DF, et al. Prediction of high-grade prostate cancer following multiparametric magnetic resonance imaging: improving the Rotterdam European randomized study of screening for prostate cancer risk calculators. Eur Urol. 2019;75:310–8.
Patel HD, Koehne EL, Shea SM, Fang AM, Gerena M, Gorbonos A, et al. A prostate biopsy risk calculator based on MRI: development and comparison of the Prospective Loyola University multiparametric MRI (PLUM) and Prostate Biopsy Collaborative Group (PBCG) risk calculators. BJU Int. 2022. https://pubmed-ncbi-nlm-nih-gov.ezproxy.galter.northwestern.edu/35733400/.
Patel HD, Koehne EL, Shea SM, Bhanji Y, Gerena M, Gorbonos A, et al. Risk of prostate cancer for men with prior negative biopsies undergoing magnetic resonance imaging compared with biopsy-naive men: a prospective evaluation of the PLUM cohort. Cancer. 2022;128:75–84. https://onlinelibrary-wiley-com.ezproxy.galter.northwestern.edu/doi/full/10.1002/cncr.33875.
Eyrich NW, Morgan TM, Tosoian JJ. Biomarkers for detection of clinically significant prostate cancer: contemporary clinical data and future directions. Transl Androl Urol. 2021;10:3091–103. https://pubmed-ncbi-nlm-nih-gov.ezproxy.galter.northwestern.edu/34430413/.
Loeb S, Shin SS, Broyles DL, Wei JT, Sanda M, Klee G, et al. Prostate Health Index (phi) improves multivariable risk prediction of aggressive prostate cancer. BJU Int. 2017;120:61.
Konety B, Zappala SM, Parekh DJ, Osterhout D, Schock J, Chudler RM, et al. The 4Kscore® test reduces prostate biopsy rates in community and academic urology practices. Rev Urol. 2015;17:231.
Fütterer JJ, Briganti A, De Visschere P, Emberton M, Giannarini G, Kirkham A, et al. Can clinically significant prostate cancer be detected with multiparametric magnetic resonance imaging? A systematic review of the literature. Eur Urol. 2015;68:1045–53.
Shariat SF, Karakiewicz PI, Suardi N, Kattan MW. Comparison of nomograms with other methods for predicting outcomes in prostate cancer: a critical analysis of the literature. Clin Cancer Res. 2008;14:4400–7.
Chun FK, Karakiewicz PI, Briganti A, Walz J, Kattan MW, Huland H, et al. A critical appraisal of logistic regression-based nomograms, artificial neural networks, classification and regression-tree models, look-up tables and risk-group stratification models for prostate cancer. BJU Int. 2007;99:794–800.
Touijer K, Scardino PT. Nomograms for staging, prognosis, and predicting treatment outcomes. Cancer. 2009;115:3107–11.
Kim SP, Karnes RJ, Nguyen PL, Ziegenfuss JY, Han LC, Thompson RH, et al. Clinical implementation of quality of life instruments and prediction tools for localized prostate cancer: results from a National Survey of Radiation Oncologists and Urologists. J Urol. 2013;189:2092–8. https://doi.org/10.1016/j.juro.2012.11.174.
Eklund M, Jäderling F, Discacciati A, Bergman M, Annerstedt M, Aly M, et al. MRI-targeted or standard biopsy in prostate cancer screening. N Engl J Med. 2021;385:908–20. https://doi.org/10.1056/NEJMoa2100852.
Nordström T, Discacciati A, Bergman M, Clements M, Aly M, Annerstedt M, et al. Prostate cancer screening using a combination of risk-prediction, MRI, and targeted prostate biopsies (STHLM3-MRI): a prospective, population-based, randomised, open-label, non-inferiority trial. Lancet Oncol. 2021;22:1240–9.
Wagaskar VG, Sobotka S, Ratnani P, Young J, Lantz A, Parekh S, et al. A 4K score/MRI-based nomogram for predicting prostate cancer, clinically significant prostate cancer, and unfavorable prostate cancer. Cancer Rep. 2021;4:1–9.
Falagario UG, Jambor I, Lantz A, Ettala O, Stabile A, Taimen P, et al. Combined use of prostate-specific antigen density and magnetic resonance imaging for prostate biopsy decision planning: a retrospective multi-institutional study using the prostate magnetic resonance imaging outcome database (PROMOD). Eur Urol Oncol. 2021;4:971–9.
Funding
Urology Care Foundation: Robert J Krane, MD Resident Research Award.
Author information
Authors and Affiliations
Contributions
MRS – patient record review, data analysis, manuscript preparation. EVL - patient record review, data analysis, manuscript preparation. SKSRK – Biostatstics. AB – patient record review. JSL – patient record review. AKM – patient record review, manuscript preparation. JAA – patient record review, data analysis. PVS – patient record review, manuscript preparation. BA – patient record review, data analytics. JMR – patient record review, data cleaning. SASM – data analytics. MKK – data support QM – data analytics support. XM – biostatistics. JJT – mentorship and guidance with the project. EMS – mentorship and guidance HDP – mentorship, guidance, and manuscript preparation. AER – mentorship, guidance with all aspects of the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Siddiqui, M.R., Li, E.V., Kumar, S.K.S.R. et al. Optimizing detection of clinically significant prostate cancer through nomograms incorporating mri, clinical features, and advanced serum biomarkers in biopsy naïve men. Prostate Cancer Prostatic Dis 26, 588–595 (2023). https://doi.org/10.1038/s41391-023-00660-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41391-023-00660-8
This article is cited by
-
Index tumor location affected early biochemical recurrence after radical prostatectomy in patients with negative surgical margin: a retrospective study
BMC Urology (2024)
-
Detection of clinically significant prostate cancer following initial omission of biopsy in multiparametric MRI era
Prostate Cancer and Prostatic Diseases (2024)
-
A novel conditional survival nomogram for monitoring real-time prognosis of non-metastatic colorectal cancer
Discover Oncology (2024)