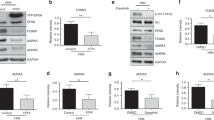
Abstract
Klotho, a 1012 amino acid transmembrane protein, is a potent tumor suppressor in different cancer types. Klotho is composed of two internal repeats KL1 and KL2, and the tumor suppressor activity is primarily attributed to the KL1 domain. Despite its significant role in regulating various cancer-related pathways, the precise mechanism underlying its tumor suppressor activity remains unresolved. In this study, we aimed to identify the sequence responsible for the tumor suppressor function of Klotho and gain insights into its mechanism of action. To accomplish this, we generated expression vectors of truncated KL1 at the C and N-terminal regions and evaluated their ability to inhibit the colony formation of several cancer cell lines. Our findings demonstrated that truncated KL1 1–340 (KL340) effectively inhibited colony formation similar to KL1, while truncated KL1 1–320 (KL320) lost this activity. Furthermore, this correlated with the inhibitory effect of KL1 and KL340 on the Wnt/β-catenin pathway, whereas KL320 had no effect. Transcriptomic analysis of MCF-7 cells expressing the constructs revealed enriched pathways associated with tumor suppressor activity in KL1 and KL340. Interestingly, the α-fold predictor tool highlighted distinct differences in the α and β sheets of the TIM barrel fold of the truncated Klotho constructs, adding to our understanding of their structural variations. In summary, this study identified the 340 N-terminal amino acids as the sequence that possesses Klotho’s tumor suppressor activity and reveals a critical role in the 320–340 sequence for this function. It also provides a foundation for the development of Klotho-based therapeutic approaches for cancer treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
RNA-seq data were deposited in the NCBI sequence read archive (SRA) under accession number PRJNA1032761.
References
Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, et al. Mutation of the mouse klotho gene leads to a syndrome resembling aging. Nature. 1997;390:45–51. https://doi.org/10.1038/36285.
Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–33.
Nabeshima YI. Klotho: a fundamental regulator of aging. Ageing Res Rev. 2002;1:627–38.
Kim J-H, Hwang K-H, Park K-S, Kong ID, Cha S-K. Biological role of anti-aging protein Klotho. J Lifestyle Med. 2015;5:1–6. https://doi.org/10.15280/jlm.2015.5.1.1.
Imura A, Iwano A, Tohyama O, Tsuji Y, Nozaki K, Hashimoto N, et al. Secreted Klotho protein in sera and CSF: implication for post-translational cleavage in release of Klotho protein from cell membrane. FEBS Lett. 2004;565:143–7.
Matsumura Y, Aizawa H, Shiraki-Iida T, Nagai R, Kuro-O M, Nabeshima YI. Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem Biophys Res Commun. 1998;242:626–30. https://doi.org/10.1006/bbrc.1997.8019.
Liu H, Fergusson MM, Castilho RM, Liu J, Cao L, Chen J, et al. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317:803–6. https://doi.org/10.1126/science.1143578.
Chen CD, Podvin S, Gillespie E, Leeman SE, Abraham CR. Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proc Natl Acad Sci USA. 2007;104:19796–801. https://doi.org/10.1073/pnas.0709805104.
Razzaque MS. The FGF23-Klotho axis: endocrine regulation of phosphate homeostasis. Nat Rev Endocrinol. 2009;5:611–9.
Chen G, Liu Y, Goetz R, Fu L, Jayaraman S, Hu MC, et al. α-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature. 2018;553:461–6. https://doi.org/10.1038/nature25451.
Abramovitz L, Rubinek T, Ligumsky H, Bose S, Barshack I, Avivi C, et al. KL1 internal repeat mediates klotho tumor suppressor activities and inhibits bFGF and IGF-I signaling in pancreatic cancer. Clin Cancer Res. 2011;17:4254–66. https://doi.org/10.1158/1078-0432.CCR-10-2749.
Cha S-K, Hu M-C, Kurosu H, Kuro-o M, Moe O, Huang C-L. Regulation of renal outer medullary potassium channel and renal K(+) excretion by Klotho. Mol Pharm. 2009;76:38–46.
Cha SK, Ortega B, Kurosu H, Rosenblatt KP, Kuro-o M, Huang CL. Removal of sialic acid involving Klotho causes cell-surface retention of TRPV5 channel via binding to galectin-1. Proc Natl Acad Sci USA. 2008;105:9805–10. https://doi.org/10.1073/pnas.0803223105.
Chang Q, Hoefs S, van der Kemp AW, Topala CN, Bindels RJ, Hoenderop JG. The beta-glucuronidase klotho hydrolyzes and activates the TRPV5 channel. Science. 2005;310:490–3.
Ligumsky H, Rubinek T, Merenbakh-Lamin K, Yeheskel A, Sertchook R, Shahmoon S, et al. Tumor suppressor activity of klotho in breast cancer is revealed by structure-function analysis. Mol Cancer Res. 2015;13:1398–407. https://doi.org/10.1158/1541-7786.MCR-15-0141.
Rubinek T, Wolf I. The role of Alpha-Klotho as a universal tumor suppressor. Vitam Horm. 2016;101:197–214. https://doi.org/10.1016/bs.vh.2016.03.001.
Imura A, Tsuji Y, Murata M, Maeda R, Kubota K, Iwano A, et al. Α-Klotho as a regulator of calcium homeostasis. Science. 2007;316:1615–8.
Shmulevich R, Nissim TB-K, Wolf I, Merenbakh-Lamin K, Fishman D, Sekler I, et al. Klotho rewires cellular metabolism of breast cancer cells through alteration of calcium shuttling and mitochondrial activity. Oncogene. 2020;39:4636–49.
Wolf I, Levanon-Cohen S, Bose S, Ligumsky H, Sredni B, Kanety H, et al. Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene. 2008;27:7094–105. https://doi.org/10.1038/onc.2008.292.
Liu F, Wu S, Ren H, Gu J. Klotho suppresses RIG-I-mediated senescence-associated inflammation. Nat Cell Biol. 2011;13:254–62. https://doi.org/10.1038/ncb2167.
Chen B, Ma X, Liu S, Zhao W, Wu J. Inhibition of lung cancer cells growth, motility and induction of apoptosis by Klotho, a novel secreted Wnt antagonist, in a dose-dependent manner. Cancer Biol Ther. 2012;13:1221–8.
Chen B, Wang X, Zhao W, Wu J. Klotho inhibits growth and promotes apoptosis in human lung cancer cell line A549. J Exp Clin Cancer Res. 2010;29:1–7.
Chen X, Tan H, Xu J, Tian Y, Yuan Q, Zuo Y, et al. Klotho-derived peptide 6 ameliorates diabetic kidney disease by targeting Wnt/β-catenin signaling. Kidney Int. 2022;102:506–20.
Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–73.
Ligumsky H, Merenbakh-Lamin K, Keren-Khadmy N, Wolf I, Rubinek T. The role of α-klotho in human cancer: molecular and clinical aspects. Oncogene. 2022;41:4487–97.
Tang X, Wang Y, Fan Z, Ji G, Wang M, Lin J, et al. Klotho: a tumor suppressor and modulator of the Wnt/β-catenin pathway in human hepatocellular carcinoma. Lab Investig. 2016;96:197–205.
Arbel Rubinstein T, Shahmoon S, Zigmond E, Etan T, Merenbakh-Lamin K, Pasmanik-Chor M, et al. Klotho suppresses colorectal cancer through modulation of the unfolded protein response. Oncogene. 2019;38:794–807.
Khorsandi L, Orazizadeh M, Niazvand F, Abbaspour MR, Mansouri E, Khodadadi A. Quercetin induces apoptosis and necroptosis in MCF-7 breast cancer cells. Bratisl Lek Listy. 2017;118:123–8.
Gong Y, Fan Z, Luo G, Yang C, Huang Q, Fan K, et al. The role of necroptosis in cancer biology and therapy. Mol Cancer. 2019;18:1–17.
Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–90.
Izquierdo MC, Perez-Gomez MV, Sanchez-Niño MD, Sanz AB, Ruiz-Andres O, Poveda J, et al. Klotho, phosphate and inflammation/ageing in chronic kidney disease. Nephrol Dial Transplant. 2012;27:6–10.
Martin M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17. https://doi.org/10.14806/ej.17.1.200.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38:276–8.
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–9.
Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21.
Yu G, Wang LG, Han Y, He QY. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9.
Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679–82.
Madeira F, Pearce M, Tivey ARN, Basutkar P, Lee J, Edbali O, et al. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022;50:W276–9.
Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:320–4.
Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:695–9.
Author information
Authors and Affiliations
Contributions
IW, TR and HL conceptualized the project. IW, TR and KM-L supervised the project. IW, TR, KM-L and MA designed the experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abboud, M., Merenbakh-Lamin, K., Volkov, H. et al. Revealing the tumor suppressive sequence within KL1 domain of the hormone Klotho. Oncogene 43, 354–362 (2024). https://doi.org/10.1038/s41388-023-02904-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02904-2