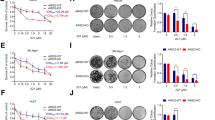
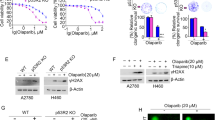
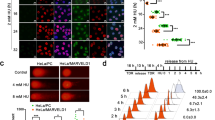
Abstract
Homologous recombination (HR) is a major DNA double-strand break (DSB) repair pathway of clinical interest because of treatment with poly(ADP-ribose) polymerase inhibitors (PARPi). Cooperation between RAD51 and BRCA2 is pivotal for DNA DSB repair, and its dysfunction induces HR deficiency and sensitizes cancer cells to PARPi. The depletion of the DEAD-box protein DDX11 was found to suppress HR in hepatocellular carcinoma (HCC) cells. The HR ability of HCC cells is not always dependent on the DDX11 level because of natural DDX11 mutations. In Huh7 cells, natural DDX11 mutations were detected, increasing the susceptibility of Huh7 cells to olaparib in vitro and in vivo. The HR deficiency of Huh7 cells was restored when CRISPR/Cas9–mediated knock-in genomic editing was used to revert the DDX11 Q238H mutation to wild type. The DDX11 Q238H mutation impeded the phosphorylation of DDX11 by ATM at serine 237, preventing the recruitment of RAD51 to damaged DNA sites by disrupting the interaction between RAD51 and BRCA2. Clinically, a high level of DDX11 correlated with advanced clinical characteristics and a poor prognosis and served as an independent risk factor for overall and disease-free survival in patients with HCC. We propose that HCC with a high level of wild-type DDX11 tends to be more resistant to PARPi because of enhanced recombination repair, and the key mutation of DDX11 (Q238H) is potentially exploitable.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data are available from the authors upon reasonable request.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Ronot M, Bouattour M, Wassermann J, Bruno O, Dreyer C, Larroque B, et al. Alternative response criteria (Choi, European association for the study of the liver, and modified Response Evaluation Criteria in Solid Tumors [RECIST]) Versus RECIST 1.1 in patients with advanced hepatocellular carcinoma treated with sorafenib. Oncologist. 2014;19:394–402.
Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–87.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7.
Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–28.
San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57.
Sasaki M, Lange J, Keeney S. Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol. 2010;11:182–95.
De Soto JA, Deng C-X. PARP-1 inhibitors: are they the long-sought genetically specific drugs for BRCA1/2-associated breast cancers? Int J Med Sci. 2006;3:117–23.
Murai J, Huang SN, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–99.
Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34.
Gerring SL, Spencer F, Hieter P. The CHL 1 (CTF 1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990;9:4347–58.
Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–49.
van der Lelij P, Chrzanowska KH, Godthelp BC, Rooimans MA, Oostra AB, Stumm M, et al. Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am J Hum Genet. 2010;86:262–6.
van Schie JJM, Faramarz A, Balk JA, Stewart GS, Cantelli E, Oostra AB, et al. Warsaw Breakage Syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion. Nat Commun. 2020;11:4287.
Jegadesan NK, Branzei D. DDX11 loss causes replication stress and pharmacologically exploitable DNA repair defects. Proc Natl Acad Sci USA. 2021;118:e2024258118.
Bhattacharya C, Wang X, Becker D. The DEAD/DEAH box helicase, DDX11, is essential for the survival of advanced melanomas. Mol Cancer. 2012;11:82.
Sui Y, Liu J, Zhang J, Zheng Z, Wang Z, Jia Z, et al. Expression and gene regulation network of adenosine receptor A2B in Lung Adenocarcinoma: a potential diagnostic and prognostic biomarker. Front Mol Biosci. 2021;8:663011.
Calì F, Bharti SK, Di Perna R, Brosh RM, Pisani FM. Tim/Timeless, a member of the replication fork protection complex, operates with the Warsaw breakage syndrome DNA helicase DDX11 in the same fork recovery pathway. Nucleic Acids Res. 2016;44:705–17.
Cortone G, Zheng G, Pensieri P, Chiappetta V, Tatè R, Malacaria E, et al. Interaction of the Warsaw breakage syndrome DNA helicase DDX11 with the replication fork-protection factor Timeless promotes sister chromatid cohesion. PLoS Genet. 2018;14:e1007622.
Lerner LK, Holzer S, Kilkenny ML, Šviković S, Murat P, Schiavone D, et al. Timeless couples G-quadruplex detection with processing by DDX11 helicase during DNA replication. EMBO J. 2020;39:e104185.
Abe T, Ooka M, Kawasumi R, Miyata K, Takata M, Hirota K, et al. Warsaw breakage syndrome DDX11 helicase acts jointly with RAD17 in the repair of bulky lesions and replication through abasic sites. Proc Natl Acad Sci USA. 2018;115:8412–7.
Stoepker C, Faramarz A, Rooimans MA, van Mil SE, Balk JA, Velleuer E, et al. DNA helicases FANCM and DDX11 are determinants of PARP inhibitor sensitivity. DNA Repair. 2015;26:54–64.
Inoue A, Li T, Roby SK, Valentine MB, Inoue M, Boyd K, et al. Loss of ChlR1 helicase in mouse causes lethality due to the accumulation of aneuploid cells generated by cohesion defects and placental malformation. Cell Cycle. 2007;6:1646–54.
Rossi F, Helbling-Leclerc A, Kawasumi R, Jegadesan NK, Xu X, Devulder P, et al. SMC5/6 acts jointly with Fanconi anemia factors to support DNA repair and genome stability. EMBO Rep. 2020;21:e48222.
Olivieri M, Cho T, Álvarez-Quilón A, Li K, Schellenberg MJ, Zimmermann M, et al. A Genetic Map of the Response to DNA Damage in Human Cells. Cell. 2020;182:481–96.e21.
Gunn A, Stark JM. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol Biol. 2012;920:379–91.
Luo K, Li L, Li Y, Wu C, Yin Y, Chen Y, et al. A phosphorylation-deubiquitination cascade regulates the BRCA2-RAD51 axis in homologous recombination. Genes Dev. 2016;30:2581–95.
Khanna KK, Lavin MF, Jackson SP, Mulhern TD. ATM, a central controller of cellular responses to DNA damage. Cell Death Differ. 2001;8:1052–65.
Kastan MB, Lim DS. The many substrates and functions of ATM. Nat Rev Mol Cell Biol. 2000;1:179–86.
Wang C, Tang H, Geng A, Dai B, Zhang H, Sun X, et al. Rational combination therapy for hepatocellular carcinoma with PARP1 and DNA-PK inhibitors. Proc Natl Acad Sci USA. 2020;117:26356–65.
Li J, Liu L, Liu X, Xu P, Hu Q, Yu Y. The role of upregulated DDX11 as a potential prognostic and diagnostic biomarker in lung adenocarcinoma. J Cancer. 2019;10:4208–16.
Hirota Y, Lahti JM. Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res. 2000;28:917–24.
Motegi A, Masutani M, Yoshioka K-I, Bessho T. Aberrations in DNA repair pathways in cancer and therapeutic significances. Semin Cancer Biol. 2019;58:29–46.
Brown JS, O’Carrigan B, Jackson SP, Yap TA. Targeting DNA repair in cancer: beyond PARP Inhibitors. Cancer Discov. 2017;7:20–37.
Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud J-B, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148.
Guo J, Duan L, He X, Li S, Wu Y, Xiang G, et al. A combined model of human iPSC-derived liver organoids and hepatocytes reveals ferroptosis in DGUOK mutant mtDNA depletion syndrome. Adv Sci. 2021;8:2004680.
Zhou Y, Caron P, Legube G, Paull TT. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014;42:e19.
Wang S, Cao K, Liao Y, Zhang W, Zheng J, Li X, et al. CDCA2 protects against oxidative stress by promoting BRCA1-NRF2 signaling in hepatocellular carcinoma. Oncogene. 2021;40:4368–83.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82002104, to KC; 81971329, to XGL), Guangdong Basic and Applied Basic Research Foundation (2019A1515110659 and 2022A1515010191), Key project of universities in Guangdong Province (2022ZDZX2023), Discipline Construction Project of Guangdong Medical University (4SG21008G), Youth Research Project of Guangdong Medical University (GDMUD2022004), Talent Development Foundation of The First Dongguan Affiliated Hospital of Guangdong Medical University (GCC2022007).
Author information
Authors and Affiliations
Contributions
KC performed most of the experiments. RNW and LHL performed mouse work and IHC analysis. YTL, XH, RXL and XWL provided technical and data analysis assistance. KC, XDX, YJW and XGL designed and interpreted the experiments and wrote the manuscript; YTL participated in proofreading this manuscript; KC, XDX, YJW and XGL revised the manuscript; XGL conceived and supervised the entire project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, K., Wang, R., Li, L. et al. Targeting DDX11 promotes PARP inhibitor sensitivity in hepatocellular carcinoma by attenuating BRCA2-RAD51 mediated homologous recombination. Oncogene 43, 35–46 (2024). https://doi.org/10.1038/s41388-023-02898-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02898-x