Abstract
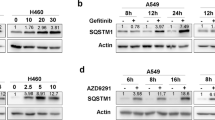
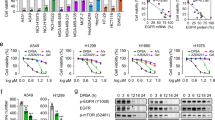
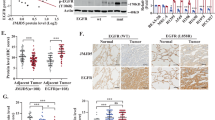
In non-small cell lung cancer (NSCLC), the overexpression or abnormal activation of epidermal growth factor receptor (EGFR) is associated with tumor progression and drug resistance. EGFR tyrosine kinase inhibitors (TKIs) are currently the first-line treatment of NSCLC. However, patients inevitably acquired EGFR TKIs resistance mutations, which led to disease progression, so it is urgent to find new treatment. Here, we report that D-mannose up-regulates lysosomal activity by enhancing TFE3-mediated lysosomal biogenesis, thereby increasing the degradation of EGFR and significantly down-regulating its protein level. Therefore, D-mannose significantly inhibited the proliferation, migration and invasion of wild-type EGFR (WT-EGFR) and EGFR mutant cells (E746-A750 deletion, L858R and T790M mutations) in vitro. Oral administration of D-mannose strongly inhibited tumor growth in mice, showing similar effects with osimertinib. Taken together, these data suggest that D-mannose may represent a new strategy for clinical treatment of NSCLC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting the findings of this study are available from the corresponding author (Lei Lv, lvlei@fudan.edu.cn) upon request.
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–74.
Sporn MB, Todaro GJ. Autocrine secretion and malignant transformation of cells. N Engl J Med. 1980;303:878–80.
Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183–232.
Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–16.
Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–54.
Mazzarella L, Guida A, Curigliano G. Cetuximab for treating non-small cell lung cancer. Expert Opin Biol Ther. 2018;18:483–93.
Ke EE, Wu YL. EGFR as a pharmacological target in EGFR-mutant non-small-cell lung cancer: where do we stand now? Trends Pharmacol Sci. 2016;37:887–903.
Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046–61.
Sharma V, Ichikawa M, Freeze HH. Mannose metabolism: more than meets the eye. Biochem Biophys Res Commun. 2014;453:220–8.
Gonzalez PS, O’Prey J, Cardaci S, Barthet VJA, Sakamaki JI, Beaumatin F, et al. Mannose impairs tumour growth and enhances chemotherapy. Nature. 2018;563:719–23.
Zhang R, Yang Y, Dong W, Lin M, He J, Zhang X, et al. D-mannose facilitates immunotherapy and radiotherapy of triple-negative breast cancer via degradation of PD-L1. Proc Natl Acad Sci USA. 2022;119:e2114851119.
Wang J, Jalali Motlagh N, Wang C, Wojtkiewicz GR, Schmidt S, Chau C. et al. d-mannose suppresses oxidative response and blocks phagocytosis in experimental neuroinflammation. Proc Natl Acad Sci USA. 2021;118:e2107663118.
Torretta S, Scagliola A, Ricci L, Mainini F, Di Marco S, Cuccovillo I, et al. D-mannose suppresses macrophage IL-1beta production. Nat Commun. 2020;11:6343.
Zhang D, Chia C, Jiao X, Jin W, Kasagi S, Wu R, et al. D-mannose induces regulatory T cells and suppresses immunopathology. Nat Med. 2017;23:1036–45.
Wang Y, Xie S, He B. Mannose shows antitumour properties against lung cancer via inhibiting proliferation, promoting cisplatin-mediated apoptosis and reducing metastasis. Mol Med Rep. 2020;22:2957–65.
Luo Q, Li B, Li G. Mannose suppresses the proliferation and metastasis of lung cancer by targeting the ERK/GSK-3β/β-catenin/SNAIL axis. OncoTargets Therapy. 2020;13:2771–81.
Yarden Y, Shilo BZ. SnapShot: EGFR signaling pathway. Cell. 2007;131:1018.
Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, Di Fiore PP. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell. 2008;15:209–19.
Perera RM, Zoncu R. The lysosome as a regulatory hub. Annu Rev Cell Dev Biol. 2016;32:223–53.
Zhang N, Dou Y, Liu L, Zhang X, Liu X, Zeng Q, et al. SA-49, a novel aloperine derivative, induces MITF-dependent lysosomal degradation of PD-L1. EBioMedicine. 2019;40:151–62.
Puertollano R, Ferguson SM, Brugarolas J, Ballabio A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 2018;37:e98804.
Ploper D, Taelman VF, Robert L, Perez BS, Titz B, Chen HW, et al. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci USA. 2015;112:E420–429.
Yin X, Wang B, Gan W, Zhuang W, Xiang Z, Han X, et al. TFE3 fusions escape from controlling of mTOR signaling pathway and accumulate in the nucleus promoting genes expression in Xp11.2 translocation renal cell carcinomas. J Exp Clin Cancer Res. 2019;38:119.
Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–8.
Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, et al. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–93.
Kruspig B, Monteverde T, Neidler S, Hock A, Kerr E, Nixon C, et al. The ERBB network facilitates KRAS-driven lung tumorigenesis. Sci Transl Med.2018;10:eaao2565.
Nukaga S, Yasuda H, Tsuchihara K, Hamamoto J, Masuzawa K, Kawada I, et al. Amplification of EGFR Wild-Type Alleles in Non-Small Cell Lung Cancer Cells Confers Acquired Resistance to Mutation-Selective EGFR Tyrosine Kinase Inhibitors. Cancer Res. 2017;77:2078–89.
Moll HP, Pranz K, Musteanu M, Grabner B, Hruschka N, Mohrherr J, et al. Afatinib restrains K-RAS-driven lung tumorigenesis. Sci Transl Med. 2018;10:eaao2301.
Yu JJ, Zhou DD, Yang XX, Cui B, Tan FW, Wang J, et al. TRIB3-EGFR interaction promotes lung cancer progression and defines a therapeutic target. Nat Commun. 2020;11:3660.
Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–10.
Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–96.
Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell. 2011;21:421–30.
Xu H, Ren D. Lysosomal physiology. Annu Rev Physiol. 2015;77:57–80.
Wang T, Zhang J, Wang S, Sun X, Wang D, Gao Y, et al. The exon 19-deleted EGFR undergoes ubiquitylation-mediated endocytic degradation via dynamin activity-dependent and -independent mechanisms. Cell Commun Signal. 2018;16:40.
Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20:3852–66.
Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–7.
Mauri V, Lotfi P, Segatori L, Sardiello M. A rapid and sensitive method for measuring N-acetylglucosaminidase activity in cultured cells. PLoS One. 2013;8:e68060.
Acknowledgements
We are grateful to members of LL laboratory for discussion throughout this study. This work was supported by the National Key R&D Program of China (2020YFA0803400/2020YFA0803402, 2022YFA0807100), the National Natural Science Foundation of China (82172936, 81972620, 82121004, 82372754, 82073128 and 32000918), Shanghai Natural Science Foundation (General Program, No. 20ZR1461900), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning and the Fundamental Research Funds for the Central Universities, “Cross” Research Fund Project of the Ninth People’s Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (JYJC202207).
Author information
Authors and Affiliations
Contributions
XS played major roles in designing and doing the experiments, analyzing the results and organizing the figures; YD, JH, HL, XY and WD helped to conduct the experiments; XX, MW, YX and LL conceived and designed the study; LL supervised the study; and XS and LL wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, X., Dai, Y., He, J. et al. D-mannose induces TFE3-dependent lysosomal degradation of EGFR and inhibits the progression of NSCLC. Oncogene 42, 3503–3513 (2023). https://doi.org/10.1038/s41388-023-02856-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02856-7