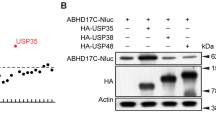
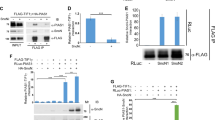
Abstract
Aberrant SUMOylation contributes to the progression of hepatocellular carcinoma (HCC), yet the molecular mechanisms have not been well elucidated. RING-type E3 ubiquitin ligase RNF146 is a key regulator of the Wnt/β-catenin signaling pathway, which is frequently hyperactivated in HCC. Here, it is identified that RNF146 can be modified by SUMO3. By mutating all lysines in RNF146, we found that K19, K61, K174 and K175 are the major sites for SUMOylation. UBC9/PIAS3/MMS21 and SENP1/2/6 mediated the conjugation and deconjugation of SUMO3, respectively. Furthermore, SUMOylation of RNF146 promoted its nuclear localization, while deSUMOylation induced its cytoplasmic localization. Importantly, SUMOylation promotes the association of RNF146 with Axin to accelerate the ubiquitination and degradation of Axin. Intriguingly, only UBC9/PIAS3 and SENP1 can act at K19/K175 in RNF146 and affect its role in regulating the stability of Axin. In addition, inhibiting RNF146 SUMOylation suppressed the progression of HCC both in vitro and in vivo. And, patients with higher expression of RNF146 and UBC9 have the worst prognosis. Taken together, we conclude that RNF146 SUMOylation at K19/K175 promotes its association with Axin and accelerates Axin degradation, thereby enhancing β-catenin signaling and contributing to cancer progression. Our findings reveal that RNF146 SUMOylation is a potential therapeutic target in HCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Albrecht LV, Tejeda-Muñoz N, De Robertis EM. Cell biology of canonical Wnt signaling. Annu Rev Cell Dev Biol. 2021;37:369–89.
Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z, et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179:561–77 e522.
Zhang Y, Chen F, Chandrashekar DS, Varambally S, Creighton CJ. Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways. Nat Commun. 2022;13:2669.
Parsons MJ, Tammela T, Dow LE. WNT as a driver and dependency in cancer. Cancer Discov. 2021;11:2413–29.
Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang X, et al. Wnt/beta-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. 2022;7:3.
Yu F, Yu C, Li F, Zuo Y, Wang Y, Yao L, et al. Wnt/beta-catenin signaling in cancers and targeted therapies. Signal Transduct Target Ther. 2021;6:307.
Han Q, Lv L, Wei J, Lei X, Lin H, Li G, et al. Vps4A mediates the localization and exosome release of beta-catenin to inhibit epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 2019;457:47–59.
Wang B, Wang T, Zhu H, Yan R, Li X, Zhang C, et al. Neddylation is essential for beta-catenin degradation in Wnt signaling pathway. Cell Rep. 2022;38:110538.
Schaefer KN, Peifer M. Wnt/beta-catenin signaling regulation and a role for biomolecular condensates. Dev Cell. 2019;48:429–44.
Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol. 2011;13:623–9.
Jackson DN, Alula KM, Delgado-Deida Y, Tabti R, Turner K, Wang X, et al. The synthetic small molecule FL3 combats intestinal tumorigenesis via Axin1-mediated inhibition of Wnt/beta-catenin signaling. Cancer Res. 2020;80:3519–29.
Fei C, Li Z, Li C, Chen Y, Chen Z, He X, et al. Smurf1-mediated Lys29-linked nonproteolytic polyubiquitination of axin negatively regulates Wnt/β-catenin signaling. Mol Cell Biol. 2013;33:4095–105.
Croy HE, Fuller CN, Giannotti J, Robinson P, Foley AVA, Yamulla RJ, et al. The poly(ADP-ribose) polymerase enzyme tankyrase antagonizes activity of the β-catenin destruction complex through ADP-ribosylation of axin and APC2. J Biol Chem. 2016;291:12747–60.
DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE, et al. Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature. 2015;517:223–6.
Morrone S, Cheng Z, Moon RT, Cong F, Xu W. Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment. Proc Natl Acad Sci USA. 2012;109:1500–5.
Han W, Koo Y, Chaieb L, Keum BR, Han JK. UCHL5 controls beta-catenin destruction complex function through Axin1 regulation. Sci Rep. 2022;12:3687.
Ji L, Lu B, Zamponi R, Charlat O, Aversa R, Yang Z, et al. USP7 inhibits Wnt/beta-catenin signaling through promoting stabilization of Axin. Nat Commun. 2019;10:4184.
Zhou L, Zheng L, Hu K, Wang X, Zhang R, Zou Y, et al. SUMOylation stabilizes hSSB1 and enhances the recruitment of NBS1 to DNA damage sites. Signal Transduct Target Ther. 2020;5:80.
Lin Q, Yu B, Wang X, Zhu S, Zhao G, Jia M, et al. K6-linked SUMOylation of BAF regulates nuclear integrity and DNA replication in mammalian cells. Proc Natl Acad Sci USA. 2020;117:10378–87.
Yang W, Robichaux WG 3rd, Mei FC, Lin W, Li L, Pan S, et al. Epac1 activation by cAMP regulates cellular SUMOylation and promotes the formation of biomolecular condensates. Sci Adv. 2022;8:eabm2960.
Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer. 2017;17:184–97.
Kumar S, Schoonderwoerd MJA, Kroonen JS, de Graaf IJ, Sluijter M, Ruano D, et al. Targeting pancreatic cancer by TAK-981: a SUMOylation inhibitor that activates the immune system and blocks cancer cell cycle progression in a preclinical model. Gut. 2022;71:2266–83.
Li J, Xu Y, Long XD, Wang W, Jiao HK, Mei Z, et al. Cbx4 governs HIF-1alpha to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell. 2014;25:118–31.
Shangguan X, He J, Ma Z, Zhang W, Ji Y, Shen K, et al. SUMOylation controls the binding of hexokinase 2 to mitochondria and protects against prostate cancer tumorigenesis. Nat Commun. 2021;12:1812.
Nakamura A, Grossman S, Song K, Xega K, Zhang Y, Cvet D, et al. The SUMOylation inhibitor subasumstat potentiates rituximab activity by IFN1-dependent macrophage and NK cell stimulation. Blood 2022;139:2770–81.
Demel UM, Boger M, Yousefian S, Grunert C, Zhang L, Hotz PW, et al. Activated SUMOylation restricts MHC class I antigen presentation to confer immune evasion in cancer. J Clin Investig. 2022;132:e152383.
Choi HK, Choi KC, Yoo JY, Song M, Ko SJ, Kim CH, et al. Reversible SUMOylation of TBL1-TBLR1 regulates β-catenin-mediated Wnt signaling. Mol Cell. 2011;43:203–16.
Karami S, Lin FM, Kumar S, Bahnassy S, Thangavel H, Quttina M, et al. Novel SUMO-protease SENP7S regulates beta-catenin signaling and mammary epithelial cell transformation. Sci Rep. 2017;7:46477.
Liu L, Sandow JJ, Leslie Pedrioli DM, Samson AL, Silke N, Kratina T, et al. Tankyrase-mediated ADP-ribosylation is a regulator of TNF-induced death. Sci Adv. 2022;8:eabh2332.
Li N, Zhang Y, Han X, Liang K, Wang J, Feng L, et al. Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumour growth. Genes Dev. 2015;29:157–70.
Hu K, Wu W, Li Y, Lin L, Chen D, Yan H, et al. Poly(ADP-ribosyl)ation of BRD7 by PARP1 confers resistance to DNA-damaging chemotherapeutic agents. EMBO Rep. 2019;20:e46166.
Shen J, Yu Z, Li N. The E3 ubiquitin ligase RNF146 promotes colorectal cancer by activating the Wnt/β-catenin pathway via ubiquitination of Axin1. Biochem Biophys Res Commun. 2018;503:991–7.
Wang H, Lu B, Castillo J, Zhang Y, Yang Z, McAllister G, et al. Tankyrase inhibitor sensitizes lung cancer cells to endothelial growth factor receptor (EGFR) inhibition via stabilizing angiomotins and inhibiting YAP signaling. J Biol Chem. 2016;291:15256–66.
Gao Y, Song C, Hui L, Li CY, Wang J, Tian Y, et al. Overexpression of RNF146 in non-small cell lung cancer enhances proliferation and invasion of tumours through the Wnt/β-catenin signaling pathway. PLoS ONE. 2014;9:e85377.
Biederstädt A, Hassan Z, Schneeweis C, Schick M, Schneider L, Muckenhuber A, et al. SUMO pathway inhibition targets an aggressive pancreatic cancer subtype. Gut. 2020;69:1472–82.
Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB, et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328:240–3.
Streich FC Jr, Lima CD. Capturing a substrate in an activated RING E3/E2-SUMO complex. Nature 2016;536:304–8.
Yunus AA, Lima CD. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol Cell. 2009;35:669–82.
Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol. 2018;36:880–7.
Zhao W, Zhang X, Rong J. SUMOylation as a therapeutic target for myocardial infarction. Front Cardiovasc Med. 2021;8:701583.
Vertegaal ACO. Signalling mechanisms and cellular functions of SUMO. Nat Rev Mol Cell Biol. 2022;23:715–31.
Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–20.
Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–47.
van Veelen W, Le NH, Helvensteijn W, Blonden L, Theeuwes M, Bakker ER, et al. β-catenin tyrosine 654 phosphorylation increases Wnt signalling and intestinal tumorigenesis. Gut. 2011;60:1204–12.
Saxena S, Zou L. Hallmarks of DNA replication stress. Mol Cell. 2022;82:2298–314.
Dagg RA, Zonderland G, Lombardi EP, Rossetti GG, Groelly FJ, Barroso S, et al. A transcription-based mechanism for oncogenic beta-catenin-induced lethality in BRCA1/2-deficient cells. Nat Commun. 2021;12:4919.
Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, et al. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014;42:W325–330.
Beauclair G, Bridier-Nahmias A, Zagury JF, Saib A, Zamborlini A. JASSA: a comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics 2015;31:3483–91.
Sharma A, Lysenko A, López Y, Dehzangi A, Sharma R, Reddy H, et al. HseSUMO: Sumoylation site prediction using half-sphere exposures of amino acids residues. BMC Genomics. 2019;19:982.
Du Y, Hou G, Zhang H, Dou J, He J, Guo Y, et al. SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 2018;46:5195–208.
Cong F, Varmus H. Nuclear-cytoplasmic shuttling of Axin regulates subcellular localization of beta-catenin. Proc Natl Acad Sci USA. 2004;101:2882–7.
Hsiao SJ, Smith S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie. 2008;90:83–92.
Chandrakumar AA, Coyaud E, Marshall CB, Ikura M, Raught B, Rottapel R. Tankyrase regulates epithelial lumen formation via suppression of Rab11 GEFs. J Cell Biol. 2021;220:e202008037.
Hu K, Li Y, Wu W, Xie L, Yan H, Cai Y, et al. ATM-dependent recruitment of BRD7 is required for transcriptional repression and DNA Repair at DNA breaks flanking transcriptional active regions. Adv Sci. 2020;7:2000157.
Varejao N, Ibars E, Lascorz J, Colomina N, Torres-Rosell J, Reverter D. DNA activates the Nse2/Mms21 SUMO E3 ligase in the Smc5/6 complex. EMBO J. 2018;37:e98306.
Kroonen JS, Vertegaal ACO. Targeting SUMO signaling to wrestle cancer. Trends Cancer. 2021;7:496–510.
Xiao J, Sun F, Wang YN, Liu B, Zhou P, Wang FX, et al. UBC9 deficiency enhances immunostimulatory macrophage activation and subsequent antitumour T cell response in prostate cancer. J Clin Investig. 2023;133:e158352.
Baik H, Boulanger M, Hosseini M, Kowalczyk J, Zaghdoudi S, Salem T, et al. Targeting the SUMO pathway primes all-trans retinoic acid-induced differentiation of nonpromyelocytic acute myeloid leukemias. Cancer Res. 2018;78:2601–13.
Kim YS, Keyser SG, Schneekloth JS Jr. Synthesis of 2’,3’,4’-trihydroxyflavone (2-D08), an inhibitor of protein sumoylation. Bioorg Med Chem Lett. 2014;24:1094–7.
Fujino N, Kubo H, Maciewicz RA. Phenotypic screening identifies Axl kinase as a negative regulator of an alveolar epithelial cell phenotype. Lab Investig. 2017;97:1047–62.
Jin Y, Nie D, Li J, Du X, Lu Y, Li Y, et al. Gas6/AXL signaling regulates self-renewal of chronic myelogenous leukemia stem cells by stabilizing β-catenin. Clin Cancer Res. 2017;23:2842–55.
Acknowledgements
This study was funded by grants from the National Key Research and Development Program of China (2021YFA0909300), the Natural Science Foundation of China (82073067, 81872140, 81621004, 81420108026 and 82172636); Guangdong Science and Technology Department (2019B020226003, 2021A0505030084, 2020B1212060018, and 2020B1212030004); Tip-top Scientific and Technical Innovative Youth Talents of Guangdong special support program (2021TQ06Y125); The Key R & D and promotion in Henan Province (212102310114).
Author information
Authors and Affiliations
Contributions
Research conception and design: DY, KSH, WJL, QFH; Experimental of methodology: KSH, WJL, YXZ, YL, WJW; Acquisition of data (provided animals, patient samples and provided facilities, etc.): QFH, JYZ, YSZ; Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): KSH, WJL, QFH, YXZ, YL, YSZ, YTQ; Write manuscript: KSH, WJL, QFH; Technical, or material support (e.g., organizing data, constructing databases,): QFH, LL, KSH.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
For the clinical specimens, ethical approval was obtained from the Ethics Committee of Sun Yat-Sen Memorial Hospital (Guangzhou, China). Informed consent was obtained from each patient. Animal study was approved by the Animal Research Committee of Sun Yat-sen University Cancer Center. Ethical approval was obtained from the Ethics Committee of Sun Yat-Sen Memorial Hospital (Guangzhou, China).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, W., Han, Q., Zhu, Y. et al. SUMOylation of RNF146 results in Axin degradation and activation of Wnt/β-catenin signaling to promote the progression of hepatocellular carcinoma. Oncogene 42, 1728–1740 (2023). https://doi.org/10.1038/s41388-023-02689-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02689-4
This article is cited by
-
Abnormal protein SUMOylation in liver disease: novel target for therapy
Journal of Molecular Medicine (2024)