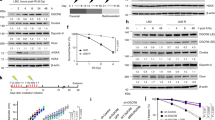
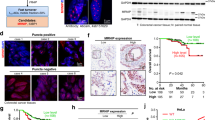
Abstract
The MRE11-RAD50-NBS1 (MRN) complex plays a crucial role in DNA double-strand breaks (DSBs) sensing and initiation of signaling cascades. However, the precise mechanisms by which the recruitment of MRN complex is regulated has yet to be elucidated. Here, we identified TRIpartite motif-containing protein 24 (TRIM24), a protein considered as an oncogene overexpressed in cancers, as a novel signaling molecule in response to DSBs. TRIM24 is essential for DSBs-induced recruitment of MRN complex and activation of downstream signaling. In the absence of TRIM24, MRN mediated DSBs repair is remarkably diminished. Mechanistically, TRIM24 is phosphorylated by ataxia-telangiectasia mutated (ATM) and then recruited to DSBs sites, facilitating the accumulation of the MRN components to chromatin. Depletion of TRIM24 sensitizes human hepatocellular carcinoma cells to cancer therapy agent-induced apoptosis and retards the tumor growth in a subcutaneous xenograft tumor mouse model. Together, our data reveal a novel function of TRIM24 in response to DSBs through regulating the MRN complex, which suggests that TRIM24 may be a potential therapeutic molecular target for tumor treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Netphos 3.1 is an online database for the prediction of kinases (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1). RNA-seq data and corresponding clinical information of HCC were extracted from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/).
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Surova O, Zhivotovsky B. Various modes of cell death induced by DNA damage. Oncogene. 2013;32:3789–97.
Huang R, Zhou PK. DNA damage repair: historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct Target Ther. 2021;6:254.
Waterman DP, Haber JE, Smolka MB. Checkpoint responses to DNA double-strand breaks. Annu Rev Biochem. 2020;89:103–33.
Trenner A, Sartori AA. Harnessing DNA double-strand break repair for cancer treatment. Front Oncol. 2019;9:1388.
Brown JS, O’Carrigan B, Jackson SP, Yap TA. Targeting DNA repair in cancer: beyond PARP inhibitors. Cancer Discov. 2017;7:20–37.
Lavin MF, Kozlov S, Gatei M, Kijas AW. ATM-dependent phosphorylation of all three members of the MRN complex: from sensor to adaptor. Biomolecules. 2015;5:2877–902.
Zhou L, Zheng L, Hu K, Wang X, Zhang R, Zou Y, et al. SUMOylation stabilizes hSSB1 and enhances the recruitment of NBS1 to DNA damage sites. Signal Transduct Target Ther. 2020;5:80.
Lee JH, Goodarzi AA, Jeggo PA, Paull TT. 53BP1 promotes ATM activity through direct interactions with the MRN complex. EMBO J. 2010;29:574–85.
Rink L, Slupianek A, Stoklosa T, Nieborowska-Skorska M, Urbanska K, Seferynska I, et al. Enhanced phosphorylation of Nbs1, a member of DNA repair/checkpoint complex Mre11-RAD50-Nbs1, can be targeted to increase the efficacy of imatinib mesylate against BCR/ABL-positive leukemia cells. Blood. 2007;110:651–60.
Zhang Y, Zhou J, Lim CU. The role of NBS1 in DNA double strand break repair, telomere stability, and cell cycle checkpoint control. Cell Res. 2006;16:45–54.
Syed A, Tainer JA. The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu Rev Biochem. 2018;87:263–94.
Bian L, Meng Y, Zhang M, Li D. MRE11-RAD50-NBS1 complex alterations and DNA damage response: implications for cancer treatment. Mol Cancer. 2019;18:169.
Wang Q, Goldstein M, Alexander P, Wakeman TP, Sun T, Feng J, et al. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. EMBO J. 2014;33:862–77.
Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, et al. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–15.
Palmer WS, Poncet-Montange G, Liu G, Petrocchi A, Reyna N, Subramanian G, et al. Structure-guided design of IACS-9571, a selective high-affinity dual TRIM24-BRPF1 bromodomain inhibitor. J Med Chem. 2016;59:1440–54.
Lv D, Li Y, Zhang W, Alvarez AA, Song L, Tang J, et al. TRIM24 is an oncogenic transcriptional co-activator of STAT3 in glioblastoma. Nat Commun. 2017;8:1454.
Chambon M, Orsetti B, Berthe ML, Bascoul-Mollevi C, Rodriguez C, Duong V, et al. Prognostic significance of TRIM24/TIF-1α gene expression in breast cancer. Am J Pathol. 2011;178:1461–9.
Liu X, Huang Y, Yang D, Li X, Liang J, Lin L, et al. Overexpression of TRIM24 is associated with the onset and progress of human hepatocellular carcinoma. PLoS ONE. 2014;9:e85462.
Groner AC, Cato L, de Tribolet-Hardy J, Bernasocchi T, Janouskova H, Melchers D, et al. TRIM24 is an oncogenic transcriptional activator in prostate cancer. Cancer Cell. 2016;29:846–58.
Bennett J, Fedorov O, Tallant C, Monteiro O, Meier J, Gamble V, et al. Discovery of a chemical tool inhibitor targeting the bromodomains of TRIM24 and BRPF. J Med Chem. 2016;59:1642–7.
Gechijian LN, Buckley DL, Lawlor MA, Reyes JM, Paulk J, Ott CJ, et al. Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat Chem Biol. 2018;14:405–12.
Jain AK, Allton K, Duncan AD, Barton MC. TRIM24 is a p53-induced E3-ubiquitin ligase that undergoes ATM-mediated phosphorylation and autodegradation during DNA damage. Mol Cell Biol. 2014;34:2695–709.
Ackerson SM, Romney C, Schuck PL, Stewart JA. To join or not to join: decision points along the pathway to double-strand break repair vs. chromosome end protection. Front Cell Dev Biol. 2021;9:708763.
Richard DJ, Savage K, Bolderson E, Cubeddu L, So S, Ghita M, et al. hSSB1 rapidly binds at the sites of DNA double-strand breaks and is required for the efficient recruitment of the MRN complex. Nucleic Acids Res. 2011;39:1692–702.
Sun X, Fu K, Hodgson A, Wier EM, Wen MG, Kamenyeva O, et al. Sam68 is required for DNA damage responses via regulating poly(ADP-ribosyl)ation. PLoS Biol. 2016;14:e1002543.
Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: to be or not to be. FEBS Lett. 2010;584:3717–24.
Huang RX, Zhou PK. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct Target Ther. 2020;5:60.
Qiu S, Huang J. MRN complex is an essential effector of DNA damage repair. J Zhejiang Univ Sci B. 2021;22:31–37.
Maréchal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5:a012716.
Kijas AW, Lim YC, Bolderson E, Cerosaletti K, Gatei M, Jakob B, et al. ATM-dependent phosphorylation of MRE11 controls extent of resection during homology directed repair by signalling through Exonuclease 1. Nucleic Acids Res. 2015;43:8352–67.
Zhou H, Kawamura K, Yanagihara H, Kobayashi J, Zhang-Akiyama QM. NBS1 is regulated by two kind of mechanisms: ATM-dependent complex formation with MRE11 and RAD50, and cell cycle-dependent degradation of protein. J Radiat Res. 2017;58:487–94.
Traven A, Heierhorst J. SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. BioEssays N Rev Mol Cell Dev Biol. 2005;27:397–407.
Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–49.
Ceccaldi R, Rondinelli B, D’Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64.
Lamarche BJ, Orazio NI, Weitzman MD. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584:3682–95.
Duursma AM, Driscoll R, Elias JE, Cimprich KA. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell. 2013;50:116–22.
Zhou Y, Paull TT. DNA-dependent protein kinase regulates DNA end resection in concert with Mre11-Rad50-Nbs1 (MRN) and ataxia telangiectasia-mutated (ATM). J Biol Chem. 2013;288:37112–25.
Limbo O, Yamada Y, Russell P. Mre11-Rad50-dependent activity of ATM/Tel1 at DNA breaks and telomeres in the absence of Nbs1. Mol Biol Cell. 2018;29:1389–99.
Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–31.
Shorstova T, Foulkes WD, Witcher M. Achieving clinical success with BET inhibitors as anti-cancer agents. Br J Cancer. 2021;124:1478–90.
Mazur PK, Herner A, Mello SS, Wirth M, Hausmann S, Sanchez-Rivera FJ, et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med. 2015;21:1163–71.
Schafer JM, Lehmann BD, Gonzalez-Ericsson PI, Marshall CB, Beeler JS, Redman LN, et al. Targeting MYCN-expressing triple-negative breast cancer with BET and MEK inhibitors. Sci Transl Med. 2020;12:eaaw8275.
Mustafi S, Camarena V, Qureshi R, Yoon H, Volmar CH, Huff TC, et al. Vitamin C supplementation expands the therapeutic window of BETi for triple negative breast cancer. EBioMedicine. 2019;43:201–10.
Burgess JT, Rose M, Boucher D, Plowman J, Molloy C, Fisher M, et al. The therapeutic potential of DNA damage repair pathways and genomic stability in lung cancer. Front Oncol. 2020;10:1256.
Rafiei S, Fitzpatrick K, Liu D, Cai MY, Elmarakeby HA, Park J, et al. ATM loss confers greater sensitivity to ATR inhibition than PARP inhibition in prostate cancer. Cancer Res. 2020;80:2094–100.
Jegadesan NK, Branzei D. DDX11 loss causes replication stress and pharmacologically exploitable DNA repair defects. Proc Natl Acad Sci USA. 2021;118:e2024258118.
Fu K, Sun X, Wier EM, Hodgson A, Liu Y, Sears CL, et al. Sam68/KHDRBS1 is critical for colon tumorigenesis by regulating genotoxic stress-induced NF-kappaB activation. eLife. 2016;5:e15018.
da Silva GN, de Camargo EA, Sávio AL, Salvadori DM. MRE11A and SKP2 genes are associated with the increased cytotoxicity induced by the synergistic effects of cisplatin and gemcitabine in bladder cancer cells. Mol Biol Rep. 2014;41:4613–21.
Araki K, Yamashita T, Reddy N, Wang H, Abuzeid WM, Khan K, et al. Molecular disruption of NBS1 with targeted gene delivery enhances chemosensitisation in head and neck cancer. Br J Cancer. 2010;103:1822–30.
Lee KC, Padget K, Curtis H, Cowell IG, Moiani D, Sondka Z, et al. MRE11 facilitates the removal of human topoisomerase II complexes from genomic DNA. Biol Open. 2012;1:863–73.
Hoa NN, Shimizu T, Zhou ZW, Wang ZQ, Deshpande RA, Paull TT, et al. Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol Cell. 2016;64:580–92.
Fu K, Sun X, Wier EM, Hodgson A, Hobbs RP, Wan F. Sam68/KHDRBS1-dependent NF-kappaB activation confers radioprotection to the colon epithelium in gamma-irradiated mice. eLife. 2016;5:e21957.
Xie J, Wen M, Zhang J, Wang Z, Wang M, Qiu Y, et al. The roles of RNA helicases in DNA damage repair and tumorigenesis reveal precision therapeutic strategies. Cancer Res. 2022;82:872–84.
Gaudreau-Lapierre A, Garneau D, Djerir B, Coulombe F, Morin T, Marechal A. Investigation of protein recruitment to DNA lesions using 405 Nm laser micro-irradiation. J Vis Exp. 2018;133:57410.
Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206.
Levone BR, Lombardi S, Barabino SML. Laser microirradiation as a tool to investigate the role of liquid-liquid phase separation in DNA damage repair. STAR Protoc. 2022;3:101146.
Euhus DM, Hudd C, LaRegina MC, Johnson FE. Tumor measurement in the nude mouse. J Surg Oncol. 1986;31:229–34.
Acknowledgements
We are grateful to Michelle C. Barton (University of Texas M. D. Anderson Cancer Center) for providing the wild-type and TRIM24 knockout MEF cells; Zhiyong Mao (Tongji University) for providing the HR reporter plasmids; the “Furong Scholar Program” provided by Department of Education of Hunan province; the Cancer Genome Atlas Program for the database.
Funding
This work was supported by National Natural Science Foundation of China [31900561, 32170726 and 32100580] to KF and YW; Hunan Provincial Science and Technology Department [2021JJ20094] to KF; Central South University [2020CX016] to KF. Funding for open access charge: National Natural Science Foundation of China.
Author information
Authors and Affiliations
Contributions
KF, FW and SX performed study concept and design; YW, YY, QW, SL, WJ and DW performed experiments and analyzed the data; YC revised the paper; JX, RT and QZ provided technical and material support. KF, YW and YY write, reviewed and revised the paper; KF provided acquisition, analysis and interpretation of data, and statistical analysis. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study is approved by the Ethics Committee of West China Hospital, Sichuan University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Yao, Y., Wei, Q. et al. TRIM24 is critical for the cellular response to DNA double-strand breaks through regulating the recruitment of MRN complex. Oncogene 42, 586–600 (2023). https://doi.org/10.1038/s41388-022-02580-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02580-8
This article is cited by
-
NPAS2 dampens chemo-sensitivity of lung adenocarcinoma cells by enhancing DNA damage repair
Cell Death & Disease (2024)