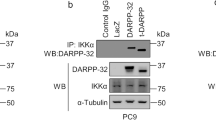
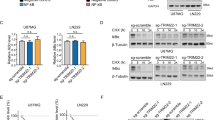
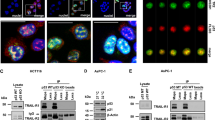
Abstract
Cellular inhibitor of apoptosis-1 (cIAP1) is a signaling regulator with oncogenic properties. It is involved in the regulation of signaling pathways controlling inflammation, cell survival, proliferation, differentiation and motility. It is recruited into membrane-receptor-associated signaling complexes thanks to the molecular adaptor TRAF2. However, the cIAP1/TRAF2 complex exists, independently of receptor engagement, in several subcellular compartments. The present work strengthens the importance of TRAF2 in the oncogenic properties of cIAP1. cIAPs-deficient mouse embryonic fibroblasts (MEFs) were transformed using the HRas-V12 oncogene. Re-expression of cIAP1 enhanced tumor growth in a nude mice xenograft model, and promoted lung tumor nodes formation. Deletion or mutation of the TRAF2-binding site completely abolished the oncogenic properties of cIAP1. Further, cIAP1 mediated the clustering of TRAF2, which was sufficient to stimulate tumor growth. Our TRAF2 interactome analysis showed that cIAP1 was critical for TRAF2 to bind to its protein partners. Thus, cIAP1 and TRAF2 would be two essential subunits of a signaling complex promoting a pro-tumoral signal. cIAP1/TRAF2 promoted the activation of the canonical NF-κB and ERK1/2 signaling pathways. NF-κB-dependent production of IL-6 triggered the activation of the JAK/STAT3 axis in an autocrine manner. Inhibition or downregulation of STAT3 specifically compromised the growth of cIAP1-restored MEFs but not that of MEFs expressing a cIAP1-mutant and treating mice with the STAT3 inhibitor niclosamide completely abrogated cIAP1/TRAF2-mediated tumor growth. Altogether, we demonstrate that cIAP1/TRAF2 binding is essential to promote tumor growth via the activation of the JAK/STAT3 signaling pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
References
Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell 1995;83:1243–52.
Hrdinka M, Yabal M. Inhibitor of apoptosis proteins in human health and disease. Genes Immun. 2019;20:641–50.
Dumetier B, Zadoroznyj A, Dubrez L IAP-mediated protein ubiquitination in regulating cell signaling. Cells 2020;9:1118.
Zadoroznyj A, Dubrez L Cytoplasmic and nuclear functions of cIAP1. Biomolecules 2022;12:322.
Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 2006;125:1253–67.
Ma O, Cai WW, Zender L, Dayaram T, Shen J, Herron AJ, et al. MMP13, Birc2 (cIAP1), and Birc3 (cIAP2), amplified on chromosome 9, collaborate with p53 deficiency in mouse osteosarcoma progression. Cancer Res. 2009;69:2559–67.
Cheng L, Zhou Z, Flesken-Nikitin A, Toshkov IA, Wang W, Camps J, et al. Rb inactivation accelerates neoplastic growth and substitutes for recurrent amplification of cIAP1, cIAP2, and Yap1 in sporadic mammary carcinoma associated with p53 deficiency. Oncogene 2010;29:5700–11.
Jin J, Xiao Y, Hu H, Zou Q, Li Y, Gao Y, et al. Proinflammatory TLR signalling is regulated by a TRAF2-dependent proteolysis mechanism in macrophages. Nat Commun. 2015;6:5930.
Yin Q, Lamothe B, Darnay BG, Wu H. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry. 2009;48:10558–67.
Dhillon B, Aleithan F, Abdul-Sater Z, Abdul-Sater AA. The evolving role of TRAFs in mediating inflammatory responses. Front Immunol. 2019;10:104.
Marivin A, Berthelet J, Cartier J, Paul C, Gemble S, Morizot A, et al. cIAP1 regulates TNF-mediated cdc42 activation and filopodia formation. Oncogene 2014;33:5534–45.
Vischioni B, Giaccone G, Span SW, Kruyt FA, Rodriguez JA. Nuclear shuttling and TRAF2-mediated retention in the cytoplasm regulate the subcellular localization of cIAP1 and cIAP2. Exp Cell Res. 2004;298:535–48.
Zhou AY, Shen RR, Kim E, Lock YJ, Xu M, Chen ZJ, et al. IKKepsilon-mediated tumorigenesis requires K63-linked polyubiquitination by a cIAP1/cIAP2/TRAF2 E3 ubiquitin ligase complex. Cell Rep. 2013;3:724–33.
Dupoux A, Cartier J, Cathelin S, Filomenko R, Solary E, Dubrez-Daloz L. cIAP1-dependent TRAF2 degradation regulates the differentiation of monocytes into macrophages and their response to CD40 ligand. Blood 2009;113:175–85.
Kreckel J, Anany MA, Siegmund D, Wajant H. TRAF2 controls death receptor-induced Caspase-8 processing and facilitates proinflammatory signaling. Front Immunol. 2019;10:2024.
Mace PD, Smits C, Vaux DL, Silke J, Day CL. Asymmetric recruitment of cIAPs by TRAF2. J Mol Biol. 2010;400:8–15.
Zheng C, Kabaleeswaran V, Wang Y, Cheng G, Wu H. Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation. Mol Cell. 2010;38:101–13.
Baud V, Liu ZG, Bennett B, Suzuki N, Xia Y, Karin M. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 1999;13:1297–308.
Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–102.
Csomos RA, Brady GF, Duckett CS. Enhanced cytoprotective effects of the inhibitor of apoptosis protein cellular IAP1 through stabilization with TRAF2. J Biol Chem. 2009;284:20531–9.
Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. 2008;9:1364–70.
Li X, Yang Y, Ashwell JD. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature 2002;416:345–7.
Matsuzawa A, Tseng PH, Vallabhapurapu S, Luo JL, Zhang W, Wang H, et al. Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science 2008;321:663–8.
Shu HB, Takeuchi M, Goeddel DV. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc Natl Acad Sci USA. 1996;93:13973–8.
Borghi A, Verstrepen L, Beyaert R. TRAF2 multitasking in TNF receptor-induced signaling to NF-κB, MAP kinases and cell death. Biochem Pharm. 2016;116:1–10.
Ceccarelli A, Di Venere A, Nicolai E, De Luca A, Minicozzi V, Rosato N, et al. TNFR-associated Factor-2 (TRAF2): Not only a trimer. Biochemistry 2015;54:6153–61.
Ye H, Park YC, Kreishman M, Kieff E, Wu H. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol Cell. 1999;4:321–30.
Park HH. Structure of TRAF family: Current understanding of receptor recognition. Front Immunol. 2018;9:1999.
Peng C, Zhu F, Wen W, Yao K, Li S, Zykova T, et al. Tumor necrosis factor receptor-associated factor family protein 2 is a key mediator of the epidermal growth factor-induced ribosomal S6 kinase 2/cAMP-responsive element-binding protein/Fos protein signaling pathway. J Biol Chem. 2012;287:25881–92.
Dubrez L, Berthelet J, Glorian V. IAP proteins as targets for drug development in oncology. OncoTargets Ther. 2013;9:1285–304.
Park MH, Hong JT. Roles of NF-κB in cancer and inflammatory diseases and their therapeutic approaches. Cells 2016;5:15.
Vu NT, Park MA, Shultz MD, Bulut GB, Ladd AC, Chalfant CE. Caspase-9b interacts directly with cIAP1 to drive agonist-independent activation of NF-κB and lung tumorigenesis. Cancer Res. 2016;76:2977–89.
Xu L, Zhu J, Hu X, Zhu H, Kim HT, LaBaer J, et al. c-IAP1 cooperates with Myc by acting as a ubiquitin ligase for Mad1. Mol Cell. 2007;28:914–22.
Li H, Fang Y, Niu C, Cao H, Mi T, Zhu H, et al. Inhibition of cIAP1 as a strategy for targeting c-MYC-driven oncogenic activity. Proc Natl Acad Sci USA. 2018;115:E9317–24.
Bishop GA, Abdul-Sater AA, Watts TH. Editorial: TRAF proteins in health and disease. Front Immunol. 2019;10:326.
Sondarva G, Kundu CN, Mehrotra S, Mishra R, Rangasamy V, Sathyanarayana P, et al. TRAF2-MLK3 interaction is essential for TNF-alpha-induced MLK3 activation. Cell Res. 2010;20:89–98.
Korchnak AC, Zhan Y, Aguilar MT, Chadee DN. Cytokine-induced activation of mixed lineage kinase 3 requires TRAF2 and TRAF6. Cell Signal. 2009;21:1620–5.
Zhao Y, Conze DB, Hanover JA, Ashwell JD. Tumor necrosis factor receptor 2 signaling induces selective c-IAP1-dependent ASK1 ubiquitination and terminates mitogen-activated protein kinase signaling. J Biol Chem. 2007;282:7777–82.
Nishitoh H, Saitoh M, Mochida Y, Takeda K, Nakano H, Rothe M, et al. ASK1 is essential for JNK/SAPK activation by TRAF2. Mol Cell. 1998;2:389–95.
Chadee DN, Yuasa T, Kyriakis JM. Direct activation of mitogen-activated protein kinase kinase kinase MEKK1 by the Ste20p homologue GCK and the adapter protein TRAF2. Mol Cell Biol. 2002;22:737–49.
Annibaldi A, Meier P. Checkpoints in TNF-induced cell death: implications in inflammation and cancer. Trends Mol Med. 2018;24:49–65.
Dogan T, Harms GS, Hekman M, Karreman C, Oberoi TK, Alnemri ES, et al. X-linked and cellular IAPs modulate the stability of C-RAF kinase and cell motility. Nat Cell Biol. 2008;10:1447–55.
Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2, and TRAF3 and the kinase NIK. Nat Immunol. 2008;9:1371–8.
Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15:234–48.
Zhu S, Jin J, Gokhale S, Lu AM, Shan H, Feng J, et al. Genetic alterations of TRAF proteins in human cancers. Front Immunol. 2018;9:2111.
Didelot C, Lanneau D, Brunet M, Bouchot A, Cartier J, Jacquel A, et al. Interaction of heat-shock protein 90 beta isoform (HSP90 beta) with cellular inhibitor of apoptosis 1 (c-IAP1) is required for cell differentiation. Cell Death Differ. 2008;15:859–66.
Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015;15:2597–601.
Acknowledgements
We thank Pauline Maes from the CLIPP proteomic platform, University of Burgundy, Philippe Hammann from the Strasbourg proteomic platform, IBMC, Valérie Saint-Gorgio from zootechny center, University of Burgundy, and Romain Aucagne from the Crigen platform, University of Burgundy. This work was supported by grants from the ‘Comités de Côte d’Or et de l’Yonne’ of the ‘Ligue Contre le Cancer’ (LD), La Ligue Nationale contre le Cancer (CG’s team), the European Union and the ‘Conseil Régional de Bourgogne’, a French Government grant managed by the French National Research Agency under the program ‘Investissements d’Avenir’ with reference ANR-11-LABX-0021, and fellowships from the ‘Ministère de l’Enseignement Supérieur et de la Recherche’ of France (to BD, AZ, JB and JA), the ‘Fondation ARC pour la Recherche sur le Cancer’ (to BD). We thank the FEDER for their financial support.
Author information
Authors and Affiliations
Contributions
BD and AZ performed most of the experiments and analyzed the data. JB initiated the project, established the tumor model, and performed some in vivo experiments. SC performed immunofluorescence analysis. JA, PB, and FC helped in performing western blots, clonogenicity assays, and in vivo experiments. CR and CP brought expertise in in vivo model and performed IV injection. CG provided scientific expertise and corrected the paper and LD conceived and supervised the project, conducted the GEPIA analysis, analyzed the data and interpreted the results, wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dumétier, B., Zadoroznyj, A., Berthelet, J. et al. cIAP1/TRAF2 interplay promotes tumor growth through the activation of STAT3. Oncogene 42, 198–208 (2023). https://doi.org/10.1038/s41388-022-02544-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02544-y
This article is cited by
-
IL-8 activates fibroblasts to promote the invasion of HNSCC cells via STAT3-MMP1
Cell Death Discovery (2024)
-
Combination of bazedoxifene with chemotherapy and SMAC-mimetics for the treatment of colorectal cancer
Cell Death & Disease (2024)
-
NAP1L1 regulates BIRC2 ubiquitination modification via E3 ubiquitin ligase UBR4 and hence determines hepatocellular carcinoma progression
Cell Death Discovery (2024)