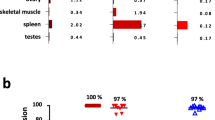
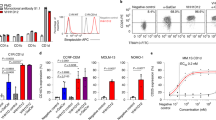
Abstract
Many therapeutic bispecific T-cell engagers (BiTEs) are in clinical trials. A modular and efficient process to create BiTEs would accelerate their development and clinical applicability. In this study, we present the design, production, and functional activity of a novel bispecific format utilizing synthetic orthogonal heterodimers to form a multichain modular design. Further addition of an immunoglobulin hinge region allowed a stable covalent linkage between the heterodimers. As proof-of-concept, we utilized CD33 and CD3 binding scFvs to engage leukemia cells and T-cells respectively. We provide evidence that this novel bispecific T-cell engager (termed IgGlue-BiTE) could bind both CD3+ and CD33+ cells and facilitates robust T-cell mediated cytotoxicity on AML cells in vitro. In a mouse model of minimal residual disease, we showed that the novel IgGlue-BiTE greatly extended survival, and mice of this treatment group were free of leukemia in the bone marrow. These findings suggest that the IgGlue-BiTE allows for robust simultaneous engagement with both antigens of interest in a manner conducive to T cell cytotoxicity against AML. These results suggest a compelling modular system for bispecific antibodies, as the CD3- and CD33-binding domains can be readily swapped with domains binding to other cancer- or immune cell-specific antigens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data relevant to the study are included in the article or uploaded as supplementary information.
References
Brinkmann U, Kontermann RE. Bispecific antibodies. Science. 2021;372:916–7.
Krah S, Sellmann C, Rhiel L, Schroter C, Dickgiesser S, Beck J, et al. Engineering bispecific antibodies with defined chain pairing. N. Biotechnol. 2017;39:167–73.
Li H, Er Saw P, Song E. Challenges and strategies for next-generation bispecific antibody-based antitumor therapeutics. Cell Mol Immunol. 2020;17:451–61.
Golay J, Choblet S, Iwaszkiewicz J, Cerutti P, Ozil A, Loisel S, et al. Design and validation of a novel generic platform for the production of tetravalent IgG1-like bispecific antibodies. J Immunol. 2016;196:3199–211.
Ha JH, Kim JE, Kim YS. Immunoglobulin Fc heterodimer platform technology: from design to applications in therapeutic antibodies and proteins. Front Immunol. 2016;7:394.
Metz S, Panke C, Haas AK, Schanzer J, Lau W, Croasdale R, et al. Bispecific antibody derivatives with restricted binding functionalities that are activated by proteolytic processing. Protein Eng Des Sel. 2012;25:571–80.
de Kruif J, Logtenberg T. Leucine zipper dimerized bivalent and bispecific scFv antibodies from a semi-synthetic antibody phage display library. J Biol Chem. 1996;271:7630–4.
Kostelny SA, Cole MS, Tso JY. Formation of a bispecific antibody by the use of leucine zippers. J Immunol. 1992;148:1547–53.
Pluckthun A, Pack P. New protein engineering approaches to multivalent and bispecific antibody fragments. Immunotechnology. 1997;3:83–105.
Chen Z, Boyken SE, Jia M, Busch F, Flores-Solis D, Bick MJ, et al. Programmable design of orthogonal protein heterodimers. Nature. 2019;565:106–11.
Payne G. Progress in immunoconjugate cancer therapeutics. Cancer Cell. 2003;3:207–12.
Molhoj M, Crommer S, Brischwein K, Rau D, Sriskandarajah M, Hoffmann P, et al. CD19-/CD3-bispecific antibody of the BiTE class is far superior to tandem diabody with respect to redirected tumor cell lysis. Mol Immunol. 2007;44:1935–43.
Roux KH, Strelets L, Michaelsen TE. Flexibility of human IgG subclasses. J Immunol. 1997;159:3372–82.
Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T et al. Protein complex prediction with AlphaFold-Multimer 2022: 2021.2010.2004.463034.
Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679–82.
Schneider D, Xiong Y, Hu P, Wu D, Chen W, Ying T, et al. A unique human immunoglobulin heavy chain variable domain-only CD33 CAR for the treatment of acute myeloid leukemia. Front Oncol. 2018;8:539.
Borot F, Wang H, Ma Y, Jafarov T, Raza A, Ali AM, et al. Gene-edited stem cells enable CD33-directed immune therapy for myeloid malignancies. Proc Natl Acad Sci USA 2019;116:11978–87.
Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82.
Acknowledgements
Research reported in this publication was performed in the CCTI Flow Cytometry Core, supported in part by the Office of the Director, National Institutes of Health under awards S10RR027050 (LSRII) and S10OD020056 (Influx sorter). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Mice imaging were performed using instrumentation maintained by the Cancer Center Small Animal Imaging Shared Resource with NIH grant #P30 CA013696 (National Cancer Institute).
Author information
Authors and Affiliations
Contributions
AB, FB, AMA, MC, and SM participated in the design of the experiments; AB, FB, MC, JD, and XD performed experiments. AB, FB, XD, JD, AMA, MC, and SM analyzed and interpreted data. AB, AMA, AMG, and SM wrote the manuscript. FB, XD, PD, MC, and JD edited the manuscript. SM conceived the idea. AMA and SM participated in the design of the study. All authors reviewed and checked the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
Columbia University has granted an exclusive option to Brahma Therapeutics to license technology that is the subject of this study. AB, FB, AMA, and SM are co-inventors on pending patent applications filed by Columbia University and optioned to Brahma Therapeutics. SM has equity ownership and is on the Scientific Advisory Board of Brahma Therapeutics. MC, JD and PD is an employee of Brahma Therapeutics.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Burke, A., Borot, F., Du, X. et al. A novel therapeutic bispecific format based on synthetic orthogonal heterodimers enables T cell activity against Acute myeloid leukemia. Oncogene 42, 26–34 (2023). https://doi.org/10.1038/s41388-022-02532-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02532-2