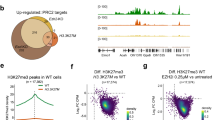
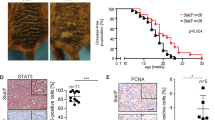
Abstract
Enhancer of Zeste Homolog 2 (EZH2) is the catalytic component of the Polycomb Repressive Complex 2, a chromatin modifying complex, which mediates methylation of lysine 27 on histone 3 (H3K27me3), a repressive chromatin mark. Genetic alterations in EZH2 in melanoma include amplifications and activating point mutations at tyrosine 641 (Y641) whose underlying oncogenic mechanisms remain largely unknown. Here, we found that expression of Ezh2Y641F causes upregulation of a subset of interferon-regulated genes in melanoma cells. Upregulation of these genes was not a direct effect of changes in H3K27me3, but via a non-canonical interaction between Ezh2 and Signal Transducer and Activator of Transcription 3 (Stat3). Ezh2 and Stat3 together function as transcriptional activators to mediate gene activation of numerous genes, including MHC Class 1b antigen processing genes. Furthermore, expression of Stat3 is required to maintain an anti-tumor immune response in Ezh2Y641F melanomas and to prevent melanoma progression and recurrence.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Marine J-C, Dawson S-J, Dawson MA. Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer. 2020;20:743–56.
Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Publ Group. 2016;17:284–99.
Network TCGA, Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, et al. Genomic classification of cutaneous melanoma. Cell 2015;161:1681–96.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data: Fig. 1. Cancer Discov. 2012;2:401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1–pl1.
Souroullas GP, Jeck WR, Parker JS, Simon JM, Liu J-Y, Paulk J, et al. An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat Med. 2016;22:632–40.
Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun. 2015;6:6051.
Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, et al. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget 2012;3:1011–25.
McHugh JB, Fullen DR, Ma L, Kleer CG, Su LD. Expression of polycomb group protein EZH2 in nevi and melanoma. J Cutan Pathol. 2007;34:597–600.
Tiffen J, Gallagher SJ, Hersey P. EZH2: an emerging role in melanoma biology and strategies for targeted therapy. Pigment Cell Melanoma Res. 2014;28:21–30.
Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. JCO. 2006;24:268–73.
Zingg D, Arenas-Ramirez N, Sahin D, Rosalia RA, Antunes AT, Haeusel J, et al. The histone methyltransferase Ezh2 controls mechanisms of adaptive resistance to tumor. Immunother Cell Rep. 2017;20:854–67.
Dasgupta M, Dermawan JKT, Willard B, Stark GR. STAT3-driven transcription depends upon the dimethylation of K49 by EZH2. Proc Natl Acad Sci USA. 2015;112:3985–90.
Kim E, Kim M, Woo D-H, Shin Y, Shin J, Chang N, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–52.
Levy DE, Darnell JE. STATs: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62.
O’Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 2002;109:S121–31.
Seidel HM, Milocco LH, Lamb P, Darnell JE, Stein RB, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. PNAS. 1995;92:3041–5.
Lerner L, Henriksen MA, Zhang X, Darnell JE. STAT3-dependent enhanceosome assembly and disassembly: synergy with GR for full transcriptional increase of the α2-macroglobulin gene. Genes Dev. 2003;17:2564–77.
Zhang X, Wrzeszczynska MH, Horvath CM, Darnell JE. Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol Cell Biol. 1999;19:7138–46.
Sun W, Snyder M, Levy DE, Zhang JJ. Regulation of Stat3 transcriptional activity by the conserved LPMSP motif for OSM and IL-6 signaling. FEBS Lett. 2006;580:5880–4.
Snyder M, Huang X-Y, Zhang JJ. Identification of novel direct Stat3 target genes for control of growth and differentiation. J Biol Chem. 2008;283:3791–8.
Tripathi SK, Chen Z, Larjo A, Kanduri K, Nousiainen K, Äijo T, et al. Genome-wide analysis of STAT3-mediated transcription during early human Th17 cell differentiation. Cell Rep. 2017;19:1888–901.
Huynh J, Chand A, Gough D, Ernst M. Therapeutically exploiting STAT3 activity in cancer — using tissue repair as a road map. Nat Rev Cancer. 2019;19:82–96.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
Zhang H, Qi J, Reyes JM, Li L, Rao PK, Li F, et al. Oncogenic deregulation of EZH2 as an opportunity for targeted therapy in lung cancer. Cancer Disco. 2016;6:1006–21.
Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA. 2010;107:21499–504.
Carpenter RL, Lo H-W. STAT3 target genes relevant to human cancers. Cancers 2014;6:897–925.
McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis -regulatory regions. Nat Biotechnol. 2010;28:495–501.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89.
Schell TD, Mylin LM, Tevethia SS, Joyce S. The assembly of functional b2-microglobulin-free MHC class I molecules that interact with peptides and CD8+ T lymphocytes. 2002;14:775–82.
Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane J-P, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun. 2017;8:1136.
Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15:234–48.
Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13:1235–42.
Burr ML, Sparbier CE, Chan KL, Chan Y-C, Kersbergen A, Lam EYN, et al. An evolutionarily conserved function of polycomb silences the MHC Class I antigen presentation pathway and enables immune evasion in cancer. Cancer Cell. 2019;36:1–26.
Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is polycomb-independent. Science 2012;338:1465–9.
Wang J, Yu X, Gong W, Liu X, Park K-S, Ma A, et al. EZH2 noncanonically binds cMyc and p300 through a cryptic transactivation domain to mediate gene activation and promote oncogenesis. Nat Cell Biol. 2022. https://doi.org/10.1038/s41556-022-00850-x.
Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci. 2009;106:5187–91.
Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–13.
Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005;436:117–22.
Kumar SM, Dai J, Li S, Yang R, Yu H, Nathanson KL, et al. Human skin neural crest progenitor cells are susceptible to BRAFV600E-induced transformation. Oncogene 2014;33:832–41.
Ballotti R, Cheli Y, Bertolotto C. The complex relationship between MITF and the immune system: a Melanoma ImmunoTherapy (response) Factor? Mol Cancer. 2020;19:170.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–48.
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6.
Furlan-Magaril M, Rincón-Arano H, Recillas-Targa F. Sequential chromatin immunoprecipitation protocol: ChIP-reChIP. Methods Mol Biol. 2009;543:253–66.
Acknowledgements
We thank the Siteman Flow Cytometry facility, McDonnel Genome Institute/Genome Access Center for technical assistance, and the Department of Comparative Medicine for animal expertise. We also thank all members of the Souroullas lab for critical input on the manuscript.
Funding
This work was supported by the US National Cancer Institute K22-CA229612-01(GPS) and T32 CA113275-10 (SZ), the Cancer Research Foundation, Chicago IL, (GPS), The Harry J. Lloyd Charitable Trust (GPS).
Author information
Authors and Affiliations
Contributions
GPS and SMZ designed experiments and wrote the manuscript. GPS, SJN and SMZ performed experiments, analyzed, and interpreted the data. LR, PYC, RLP, SS and PNL performed experiments. GPS conceived of and supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zimmerman, S.M., Nixon, S.J., Chen, P.Y. et al. Ezh2Y641F mutations co-operate with Stat3 to regulate MHC class I antigen processing and alter the tumor immune response in melanoma. Oncogene 41, 4983–4993 (2022). https://doi.org/10.1038/s41388-022-02492-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02492-7
This article is cited by
-
Cellular senescence-related genes: predicting prognosis in hepatocellular carcinoma
BMC Cancer (2023)
-
Harnessing epithelial-mesenchymal plasticity to boost cancer immunotherapy
Cellular & Molecular Immunology (2023)
-
Epigenetics behind tumor immunology: a mini review
Oncogene (2023)