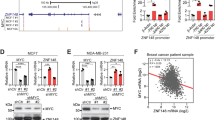
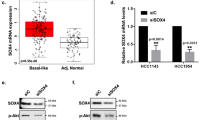
Abstract
SMARCE1 gene, encoding a core subunit of SWI/SNF chromatin remodeling complex, is situated on chromosome 17q21-ter region that is frequently gained in neuroblastoma. However, its role in the tumorigenesis remains unknown. Here, we showed that high expression of SMARCE1 was associated with poor prognosis of patients with neuroblastoma, especially those with MYCN amplification. Knockdown of SMARCE1 reduced proliferation, colony formation, and tumorigenicity of neuroblastoma cells. Mechanistically, SMARCE1 directly interacted with MYCN, which was necessary for MYCN-mediated transcriptional activation of downstream target genes including PLK1, ODC1, and E2F2. Overexpression of PLK1, ODC1 or E2F2 significantly reversed the inhibiting effect of SMARCE1 knockdown on the proliferation, colony formation, and tumorigenicity of MYCN-amplified neuroblastoma cells. Moreover, we revealed that MYCN directly regulated SMARCE1 transcription through binding to a non-canonical E-box of SMARCE1 promoter, thus enhancing SMARCE1-MYCN cooperativity. These findings establish SMARCE1 is a critical oncogenic factor in neuroblastoma and provide a new potential target for treatment of neuroblastoma with 17q21-ter gain and MYCN amplification.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All of the data and material in this paper are available when requested. We obtained several other publicly available ChIP-seq datasets (GSM2113542, GSM2214113, GSM2113529, GSM2113526 and ENCSR157TCS) for analysis. We also obtained 88 human Neuroblastoma samples dataset (GSE16476) for analysis (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi; https://www.encodeproject.org/).
References
Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, et al. Neuroblastoma. Nat Rev Dis Prim. 2016;2:16078.
Fetahu IS, Taschner-Mandl S. Neuroblastoma and the epigenome. Cancer Metastasis Rev. 2021;40:173–89.
Bown N, Cotterill S, Lastowska M, O’Neill S, Pearson AD, Plantaz D, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340:1954–61.
O’Neill S, Ekstrom L, Lastowska M, Roberts P, Brodeur GM, Kees UR, et al. MYCN amplification and 17q in neuroblastoma: evidence for structural association. Genes Chromosomes Cancer. 2001;30:87–90.
Bell JL, Turlapati R, Liu T, Schulte JH, Huttelmaier S. IGF2BP1 harbors prognostic significance by gene gain and diverse expression in neuroblastoma. J Clin Oncol. 2015;33:1285–93.
Islam A, Kageyama H, Takada N, Kawamoto T, Takayasu H, Isogai E, et al. High expression of Survivin, mapped to 17q25, is significantly associated with poor prognostic factors and promotes cell survival in human neuroblastoma. Oncogene. 2000;19:617–23.
Wong M, Sun Y, Xi Z, Milazzo G, Poulos RC, Bartenhagen C, et al. JMJD6 is a tumorigenic factor and therapeutic target in neuroblastoma. Nat Commun. 2019;10:3319.
Nagy Z, Seneviratne JA, Kanikevich M, Chang W, Mayoh C, Venkat P, et al. An ALYREF-MYCN coactivator complex drives neuroblastoma tumorigenesis through effects on USP3 and MYCN stability. Nat Commun. 2021;12:1881.
Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Bio. 2017;18:407–22.
Tsukiyama T. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat Rev Mol Cell Bio. 2002;3:422–9.
Wattler F, Wattler S, Kelly M, Skinner HB, Nehls M. Cloning, chromosomal location, and expression analysis of murine Smarce1-related, a new member of the high-mobility 365 group gene family. Genomics. 1999;60:172–8.
Heo Y, Park JH, Kim J, Han J, Yun JH, Lee W. Crystal structure of the HMG domain of human BAF57 and its interaction with four-way junction DNA. Biochem Biophys Res Commun. 2020;533:919–24.
Sethuraman A, Brown M, Seagroves TN, Wu ZH, Pfeffer LM, Fan M. SMARCE1 regulates metastatic potential of breast cancer cells through the HIF1A/PTK2 pathway. Breast Cancer Res. 2016;18:81.
Wan M, Zhang J, Lai D, Jani A, Prestone-Hurlburt P, Zhao L, et al. Molecular basis of CD4 repression by the Swi/Snf-like BAF chromatin remodeling complex. Eur J Immunol. 2009;39:580–8.
Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–16.
Monclair T, Brodeur GM, Ambros PF, Brisse HJ, Cecchetto G, Holmes K, et al. The International Neuroblastoma Risk Group (INRG) staging system: an INRG Task Force report. J Clin Oncol. 2009;27:298–303.
Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–90.
Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med. 2013;3:a014415.
Baluapuri A, Wolf E, Eilers M. Target gene-independent functions of MYC oncoproteins. Nat Rev Mol Cell Biol. 2020;21:255–67.
Wenzel A, Schwab M. The mycN/max protein complex in neuroblastoma. Short Rev Eur J Cancer. 1995;31A:516–9.
Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–70.
Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–84.
Cole KA, Huggins J, Laquaglia M, Hulderman CE, Russell MR, Bosse K, et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci USA 2011;108:3336–41.
Kocak H, Ackermann S, Hero B, Kahlert Y, Oberthuer A, Juraeva D, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. 2013;4:e586.
Bourdeaut F, Janoueix-Lerosey I, Lucchesi C, Paris R, Ribeiro A, de Pontual L, et al. Cholinergic switch associated with morphological differentiation in neuroblastoma. J Pathol. 2009;219:463–72.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Zhang W, Yu Y, Hertwig F, Thierry-Mieg J, Zhang W, Thierry-Mieg D, et al. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. 2015;16:133.
Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–93.
Krstic A, Konietzny A, Halasz M, Cain P, Oppermann U, Kolch W, et al. A chemo-genomic approach identifies diverse epigenetic therapeutic vulnerabilities in MYCN-amplified neuroblastoma. Front Cell Dev Biol. 2021;9:612518.
Duffy DJ, Konietzny A, Krstic A, Mehta JP, Halasz M, Kolch W. Identification of a MYCN and Wnt-related VANGL2-ITLN1 fusion gene in neuroblastoma. Gene Rep. 2018;12:187–200.
Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–66.
Gryder BE, Yohe ME, Chou HC, Zhang X, Marques J, Wachtel M, et al. PAX3-FOXO1 establishes myogenic super enhancers and confers BET bromodomain vulnerability. Cancer Disco. 2017;7:884–99.
Zeid R, Lawlor MA, Poon E, Reyes JM, Fulciniti M, Lopez MA, et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat Genet. 2018;50:515–23.
Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–5.
Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447.
Nakayama RT, Pulice JL, Valencia AM, McBride MJ, McKenzie ZM, Gillespie MA, et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat Genet. 2017;49:1613–23.
Sokol ES, Feng YX, Jin DX, Tizabi MD, Miller DH, Cohen MA, et al. SMARCE1 is required for the invasive progression of in situ cancers. Proc Natl Acad Sci USA 2017;114:4153–8.
Molenaar JJ, Domingo-Fernandez R, Ebus ME, Lindner S, Koster J, Drabek K, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet. 2012;44:1199–206.
Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–801.
Vervoorts J, Luscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, et al. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4:484–90.
Rust K, Tiwari MD, Mishra VK, Grawe F, Wodarz A. Myc and the Tip60 chromatin remodeling complex control neuroblast maintenance and polarity in Drosophila. Embo J. 2018;37:e98659.
Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–51.
Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–17.
Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205.
Cowling VH, Chandriani S, Whitfield ML, Cole MD. A conserved Myc protein domain, MBIV, regulates DNA binding, apoptosis, transformation, and G2 arrest. Mol Cell Biol. 2006;26:4226–39.
Amente S, Milazzo G, Sorrentino MC, Ambrosio S, Di Palo G, Lania L, et al. Lysine-specific demethylase (LSD1/KDM1A) and MYCN cooperatively repress tumor suppressor genes in neuroblastoma. Oncotarget. 2015;6:14572–83.
Kumar P, Mistri TK. Transcription factors in SOX family: potent regulators for cancer initiation and development in the human body. Semin Cancer Biol. 2020;67:105–13.
Amente S, Zhang J, Lavadera ML, Lania L, Avvedimento EV, Majello B. Myc and PI3K/AKT signaling cooperatively repress FOXO3a-dependent PUMA and GADD45a gene expression. Nucleic Acids Res. 2011;39:9498–507.
Xiao D, Yue M, Su H, Ren P, Jiang J, Li F, et al. Polo-like Kinase-1 regulates Myc stabilization and activates a feedforward circuit promoting tumor cell survival. Mol Cell. 2016;64:493–506.
Hogarty MD, Norris MD, Davis K, Liu X, Evageliou NF, Hayes CS, et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 2008;68:9735–45.
Wong M, Tee AEL, Milazzo G, Bell JL, Poulos RC, Atmadibrata B, et al. The histone methyltransferase DOT1L promotes neuroblastoma by regulating gene transcription. Cancer Res. 2017;77:2522–33.
Sun J, Yi S, Qiu L, Fu W, Wang A, Liu F, et al. SATB1 defines a subtype of cutaneous CD30(+) lymphoproliferative disorders associated with a T-Helper 17 cytokine profile. J Invest Dermatol. 2018;138:1795–804.
Wei J, Yin Y, Deng Q, Zhou J, Wang Y, Yin G, et al. Integrative analysis of MicroRNA and gene interactions for revealing candidate signatures in prostate cancer. Front Genet. 2020;11:176.
Mastronikolis N, Ragos V, Kyrodimos E, Chrysovergis A, Papanikolaou V, Mastronikolis S, et al. Mechanisms of C-myc oncogenic activity in head and neck squamous cell carcinoma. J BUON. 2019;24:2242–4.
Lee WH, Murphree AL, Benedict WF. Expression and amplification of the N-myc gene in primary retinoblastoma. Nature. 1984;309:458–60.
Hui AB, Lo KW, Yin XL, Poon WS, Ng HK. Detection of multiple gene amplifications in glioblastoma multiforme using array-based comparative genomic hybridization. Lab Invest. 2001;81:717–23.
Dias P, Kumar P, Marsden HB, Gattamaneni HR, Heighway J, Kumar S. N-myc gene is amplified in alveolar rhabdomyosarcomas (RMS) but not in embryonal RMS. Int J Cancer. 1990;45:593–6.
Nisen PD, Zimmerman KA, Cotter SV, Gilbert F, Alt FW. Enhanced expression of the N-myc gene in Wilms’ tumors. Cancer Res. 1986;46:6217–22.
Tonelli R, Purgato S, Camerin C, Fronza R, Bologna F, Alboresi S, et al. Anti-gene peptide nucleic acid specifically inhibits MYCN expression in human neuroblastoma cells leading to cell growth inhibition and apoptosis. Mol Cancer Ther. 2005;4:779–86.
Burkhart CA, Cheng AJ, Madafiglio J, Kavallaris M, Mili M, Marshall GM, et al. Effects of MYCN antisense oligonucleotide administration on tumorigenesis in a murine model of neuroblastoma. J Natl Cancer Inst. 2003;95:1394–403.
Otto T, Horn S, Brockmann M, Eilers U, Schuttrumpf L, Popov N, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78.
Albihn A, Johnsen JI, Henriksson MA. MYC in oncogenesis and as a target for cancer therapies. Adv Cancer Res. 2010;107:163–224.
Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–30.
Zafar A, Wang W, Liu G, Wang X, Xian W, McKeon F, et al. Molecular targeting therapies for neuroblastoma: progress and challenges. Med Res Rev. 2021;41:961–1021.
Bell E, Chen L, Liu T, Marshall GM, Lunec J, Tweddle DA. MYCN oncoprotein targets and their therapeutic potential. Cancer Lett. 2010;293:144–57.
Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5.
Mao L, Ding J, Zha Y, Yang L, McCarthy BA, King W, et al. HOXC9 links cell-cycle exit and neuronal differentiation and is a prognostic marker in neuroblastoma. Cancer Res. 2011;71:4314–24.
Peng W, Shi S, Zhong J, Liang H, Hou J, Hu X, et al. CBX3 accelerates the malignant progression of glioblastoma multiforme by stabilizing EGFR expression. Oncogene. 2022;41:3051–63.
Zhang K, Fu G, Pan G, Li C, Shen L, Hu R, et al. Demethylzeylasteral inhibits glioma growth by regulating the miR-30e-5p/MYBL2 axis. Cell Death Dis. 2018;9:1035.
Hou J, Deng Q, Zhou J, Zou J, Zhang Y, Tan P, et al. CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR. Oncogene. 2017;36:1134–44.
Hu X, Pei D, Ci M, Zhang G, Li B, Wang J, et al. ACTL6A deficiency induces apoptosis through impairing DNA replication and inhibiting the ATR-Chk1 signaling in glioblastoma cells. Biochem Biophys Res Commun. 2022;599:148–55.
Yang R, Li X, Wu Y, Zhang G, Liu X, Li Y, et al. EGFR activates GDH1 transcription to promote glutamine metabolism through MEK/ERK/ELK1 pathway in glioblastoma. Oncogene. 2020;39:2975–86.
Lee TH, Tun-Kyi A, Shi R, Lim J, Soohoo C, Finn G, et al. Essential role of Pin1 in the regulation of TRF1 stability and telomere maintenance. Nat Cell Biol. 2009;11:97–105.
Li Y, Su Y, Zhao Y, Hu X, Zhao G, He J, et al. Demethylzeylasteral inhibits proliferation, migration, and invasion through FBXW7/c-Myc axis in gastric cancer. MedComm. 2021;2:467–80.
Acknowledgements
This research was supported by the Natural Science Foundation of Chongqing (cstc2019jcyj-zdxmX0033, cstc2022ycjh-bgzxm0145), the pilot program of Southwest University (SWU-XDZD22006).
Author information
Authors and Affiliations
Contributions
HC, PL and XH conceived and designed the experiments. XH, JH, WP, SW, YL, GZ and XZ performed the experiments, collected the data. XH, RL and MX analyzed the data and prepared the figures. XH and HC wrote the manuscript. All the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All experiments involving cancer patients’ samples were obtained from Chaoying Biotechnology Co., Ltd. (Henan, China), and the studies were approved by the Medical Ethics Committee of Tongxu County People’s Hospital of Henan Province. All of the patients were informed consent.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41388_2022_2428_MOESM4_ESM.jpg
Supplementary Figure 2. SMARCE1 is required for the colony-formation ability and proper cell cycle progression of MYCN-amplified neuroblastoma cells
41388_2022_2428_MOESM8_ESM.jpg
Supplementary Figure 6. SMARCE1 cooperate with MYCN to transcriptionally regulates the expression of PLK1, ODC1 and E2F2
41388_2022_2428_MOESM9_ESM.jpg
Supplementary Figure 7. The MYCN-SMARCE1 interaction is an important factor that regulates PLK1 expression and cell proliferation.
41388_2022_2428_MOESM10_ESM.jpg
Supplementary Figure 8. Analysis of the correlation between MYCN and SMARCE1 expression levels in several MYCN-amplified or overexpressed cancers
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, X., Liu, R., Hou, J. et al. SMARCE1 promotes neuroblastoma tumorigenesis through assisting MYCN-mediated transcriptional activation. Oncogene 41, 4295–4306 (2022). https://doi.org/10.1038/s41388-022-02428-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02428-1