Abstract
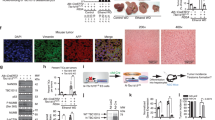
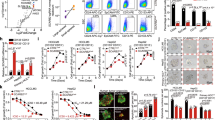
HFE (Hemochromatosis) is a conventional iron level regulator and its loss of function due to gene mutations increases the risk of cancers including hepatocellular carcinoma (HCC). Likewise, studies focusing on HFE overexpression in cancers are all limited to linking up these events as a consequence of iron level deregulation. No study has explored any iron unrelated role of HFE in cancers. Here, we first reported HFE as an oncogene in HCC and its undescribed function on promoting abscission in cytokinesis during mitotic cell division, independent of its iron-regulating ability. Clinical analyses revealed HFE upregulation in tumors linking to large tumor size and poor prognosis. Functionally and mechanistically, HFE promoted cytokinetic abscission via facilitating ESCRT abscission machinery recruitment to the abscission site through signaling a novel HFE/ALK3/Smads/LIF/Hippo/YAP/YY1/KIF13A axis. Pharmacological blockage of HFE signaling axis impeded tumor phenotypes in vitro and in vivo. Our data on HFE-driven HCC unveiled a new mechanism utilized by cancer cells to propel rapid cell division. This study also laid the groundwork for tumor intolerable therapeutics development given the high cytokinetic dependency of cancer cells and their vulnerability to cytokinetic blockage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Lens SMA, Medema RH. Cytokinesis defects and cancer. Nat Rev Cancer. 2019;19:32–45.
D’Avino PP, Giansanti MG, Petronczki M. Cytokinesis in animal cells. Cold Spring Harb Perspect Biol. 2015;7:a015834.
Donne R, Saroul-Ainama M, Cordier P, Celton-Morizur S, Desdouets C. Polyploidy in liver development, homeostasis and disease. Nat Rev Gastroenterol Hepatol. 2020;17:391–405.
Fededa JP, Gerlich DW. Molecular control of animal cell cytokinesis. Nat Cell Biol. 2012;14:440–7.
Zhang S, Nguyen LH, Zhou K, Tu HC, Sehgal A, Nassour I, et al. Knockdown of Anillin Actin Binding Protein Blocks Cytokinesis in Hepatocytes and Reduces Liver Tumor Development in Mice Without Affecting Regeneration. Gastroenterology. 2018;154:1421–34.
Xing T, Yan T, Zhou Q. Identification of key candidate genes and pathways in hepatocellular carcinoma by integrated bioinformatical analysis. Exp Ther Med. 2018;15:4932–42.
Dong F, Yang Q, Wu Z, Hu X, Shi D, Feng M, et al. Identification of survival-related predictors in hepatocellular carcinoma through integrated genomic, transcriptomic, and proteomic analyses. Biomed Pharmacother. 2019;114:108856.
Barton JC, Edwards CQ, Acton RT. HFE gene: Structure, function, mutations, and associated iron abnormalities. Gene. 2015;574:179–92.
Reuben A, Chung JW, Lapointe R, Santos MM. The hemochromatosis protein HFE 20 years later: An emerging role in antigen presentation and in the immune system. Immun Inflamm Dis. 2017;5:218–32.
Weston C, Connor J. Evidence for the Influence of the Iron Regulatory MHC Class I Molecule HFE on Tumor Progression in Experimental Models and Clinical Populations. Transl Oncogenomics. 2014;6:1–12.
Lenarduzzi M, Hui AB, Yue S, Ito E, Shi W, Williams J, et al. Hemochromatosis enhances tumor progression via upregulation of intracellular iron in head and neck cancer. PLoS One. 2013;8:e74075.
Josson S, Matsuoka Y, Gururajan M, Nomura T, Huang WC, Yang X, et al. Inhibition of beta2-microglobulin/hemochromatosis enhances radiation sensitivity by induction of iron overload in prostate cancer cells. PLoS One. 2013;8:e68366.
Wang MT, Fer N, Galeas J, Collisson EA, Kim SE, Sharib J, et al. Blockade of leukemia inhibitory factor as a therapeutic approach to KRAS driven pancreatic cancer. Nat Commun. 2019;10:3055.
Muckenthaler MU, Rivella S, Hentze MW, Galy B. A Red Carpet for Iron Metabolism. Cell. 2017;168:344–61.
Poli M, Asperti M, Ruzzenenti P, Regoni M, Arosio P. Hepcidin antagonists for potential treatments of disorders with hepcidin excess. Front Pharm. 2014;5:86.
Penuelas S, Anido J, Prieto-Sanchez RM, Folch G, Barba I, Cuartas I, et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–27.
Sagona AP, Nezis IP, Pedersen NM, Liestol K, Poulton J, Rusten TE, et al. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol. 2010;12:362–71.
Totaro A, Panciera T, Piccolo S. YAP/TAZ upstream signals and downstream responses. Nat Cell Biol. 2018;20:888–99.
Verheul TCJ, van Hijfte L, Perenthaler E, Barakat TS. The Why of YY1: Mechanisms of Transcriptional Regulation by Yin Yang 1. Front Cell Dev Biol. 2020;8:592164.
Hoxha S, Shepard A, Troutman S, Diao H, Doherty JR, Janiszewska M, et al. YAP-Mediated Recruitment of YY1 and EZH2 Represses Transcription of Key Cell-Cycle Regulators. Cancer Res. 2020;80:2512–22.
Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A. The discovery of the new haemochromatosis gene. 1996. J Hepatol. 2003;38:704–9.
Yang XM, Cao XY, He P, Li J, Feng MX, Zhang YL, et al. Overexpression of Rac GTPase Activating Protein 1 Contributes to Proliferation of Cancer Cells by Reducing Hippo Signaling to Promote Cytokinesis. Gastroenterology. 2018;155:1233–49 e22.
Tang H, Zhao H, Yu ZY, Feng X, Fu BS, Qiu CH, et al. MicroRNA-194 inhibits cell invasion and migration in hepatocellular carcinoma through PRC1-mediated inhibition of Wnt/beta-catenin signaling pathway. Dig Liver Dis. 2019;51:1314–22.
Shen H, Wang Z, Ren S, Wang W, Duan L, Zhu D, et al. Prognostic biomarker MITD1 and its correlation with immune infiltrates in hepatocellular carcinoma (HCC). Int Immunopharmacol. 2020;81:106222.
Fan Y, Du Z, Ding Q, Zhang J, Op Den Winkel M, Gerbes AL, et al. SEPT6 drives hepatocellular carcinoma cell proliferation, migration and invasion via the Hippo/YAP signaling pathway. Int J Oncol. 2021;58:25.
Maugeri-Sacca M, De Maria R. The Hippo pathway in normal development and cancer. Pharm Ther. 2018;186:60–72.
Bui DA, Lee W, White AE, Harper JW, Schackmann RC, Overholtzer M, et al. Cytokinesis involves a nontranscriptional function of the Hippo pathway effector YAP. Sci Signal. 2016;9:ra23.
Henriques AC, Ribeiro D, Pedrosa J, Sarmento B, Silva PMA, Bousbaa H. Mitosis inhibitors in anticancer therapy: When blocking the exit becomes a solution. Cancer Lett. 2019;440-441:64–81.
Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–96.
Xu H, Choe C, Shin SH, Park SW, Kim HS, Jung SH, et al. Silencing of KIF14 interferes with cell cycle progression and cytokinesis by blocking the p27(Kip1) ubiquitination pathway in hepatocellular carcinoma. Exp Mol Med. 2014;46:e97.
Yang T, Zhang XB, Zheng ZM. Suppression of KIF14 expression inhibits hepatocellular carcinoma progression and predicts favorable outcome. Cancer Sci. 2013;104:552–7.
Sun X, Jin Z, Song X, Wang J, Li Y, Qian X, et al. Evaluation of KIF23 variant 1 expression and relevance as a novel prognostic factor in patients with hepatocellular carcinoma. BMC Cancer. 2015;15:961.
Lu M, Huang X, Chen Y, Fu Y, Xu C, Xiang W, et al. Aberrant KIF20A expression might independently predict poor overall survival and recurrence-free survival of hepatocellular carcinoma. IUBMB Life. 2018;70:328–35.
Liu X, Li Y, Zhang X, Liu XY, Peng A, Chen Y, et al. Inhibition of kinesin family member 20B sensitizes hepatocellular carcinoma cell to microtubule-targeting agents by blocking cytokinesis. Cancer Sci. 2018;109:3450–60.
Acknowledgements
We would like to thank Dr Lui Ng from Department of Surgery, The University of Hong Kong for providing anti-Smad2/3 antibodies.
Funding
This study was financially supported by Theme-based Research Scheme (T42-103/16-N and T12-703/19-R) of the Research Grants Council, Hong Kong and the Health@InnoHK program of the Innovation and Technology Commission of the Hong Kong SAR Government.
Author information
Authors and Affiliations
Contributions
PD, ZC, BL, and NPL: study concept and design; PD, ZC, AKC, MWC, CHA: acquisition of data; PD, ZC, YZ, AKC, MWC, CHA: analysis and interpretation of data; PD, ZC, AKC, MWC, NPL: drafting of the manuscript; CHA, KM, NPL: critical revision of the manuscript for important intellectual content; PD, ZC, CHA: statistical analysis; CML, KM, NPL: obtained funding; AKC, MWC, CHA, WKS, TTC, KM, NPL: administrative, technical, or material support; WKS, TTC, CML, KM, NPL: study supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Dong, P., Cai, Z., Li, B. et al. HFE promotes mitotic cell division through recruitment of cytokinetic abscission machinery in hepatocellular carcinoma. Oncogene 41, 4185–4199 (2022). https://doi.org/10.1038/s41388-022-02419-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02419-2