Abstract
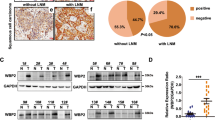
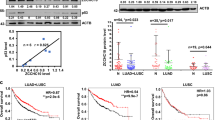
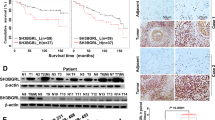
Mutant p53 (mtp53) can exert cancer-promoting activities via “gain-of-function”, which has become a popular research target. Although lots of researchers focus on the tumor-suppressor role for p53, the regulation of mutant p53 remains unknown. Here, we report a mechanism by which mtp53 regulate the transcription of Rab coupling protein (RCP) to influence lung cancer behavior. First, we show that RCP is specifically expressed at high levels in lung cancer tissues and cells, and RCP knockout suppresses tumor growth and metastasis. Further mass spectrometry and functional analysis identify that Sp1, Sp3 and Stat3 are the transcriptional activators of RCP. Moreover, p53 is involved in modulating RCP expression in an Sp1/3 dependent manner. Mechanistically, in contrast to wild-type p53 suppression of RCP transcription by decreasing Sp1/3 proteins, TP53 mutations have changed on Sp1/3 expression via “loss-of-function”. Surprisingly, the DNA contact mutants of p53 further robustly enhance their binding ability with Sp1/3 to drive RCP expression through the “gain-of-function” activity. Collectively, we reveal a mechanism by which p53 regulating the transcription of RCP to influence lung cancer progression, which provides new insights for treating p53 mutant lung cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386.
van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–70.
Kurppa KJ, Liu Y, To C, Zhang T, Fan M, Vajdi A, et al. Treatment-induced tumor dormancy through YAP-mediated transcriptional reprogramming of the apoptotic pathway. Cancer Cell. 2020;37:104–22 e112.
Prekeris R, Klumperman J, Scheller RH. A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol Cell. 2000;6:1437–48.
Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J, et al. RCP is a human breast cancer-promoting gene with Ras-activating function. J Clin Invest. 2009;119:2171–83.
Cho KH, Lee HY. Rab25 and RCP in cancer progression. Arch Pharm Res. 2019;42:101–12.
Dai Y, Liu Y, Huang D, Yu C, Cai G, Pi L, et al. Increased expression of Rab coupling protein in squamous cell carcinoma of the head and neck and its clinical significance. Oncol Lett. 2012;3:1231–6.
Mills GB, Jurisica I, Yarden Y, Norman JC. Genomic amplicons target vesicle recycling in breast cancer. J Clin Invest. 2009;119:2123–7.
Balsara BR, Sonoda G, du Manoir S, Siegfried JM, Gabrielson E, Testa JR. Comparative genomic hybridization analysis detects frequent, often high-level, overrepresentation of DNA sequences at 3q, 5p, 7p, and 8q in human non-small cell lung carcinomas. Cancer Res. 1997;57:2116–20.
Tang Q, Lento A, Suzuki K, Efe G, Karakasheva T, Long A, et al. Rab11-FIP1 mediates epithelial-mesenchymal transition and invasion in esophageal cancer. EMBO Rep. 2021;22:e48351.
Phatak V, von Grabowiecki Y, Janus J, Officer L, Behan C, Aschauer L, et al. Mutant p53 promotes RCP-dependent chemoresistance coinciding with increased delivery of P-glycoprotein to the plasma membrane. Cell Death Dis. 2021;12:207.
Zhang S, Wang C, Ma B, Xu M, Xu S, Liu J, et al. Mutant p53 Drives Cancer Metastasis via RCP-Mediated Hsp90alpha Secretion. Cell Rep. 2020;32:107879.
Kim JY, Cho KH, Jeong BY, Park CG, Lee HY. Zeb1 for RCP-induced oral cancer cell invasion and its suppression by resveratrol. Exp Mol Med. 2020;52:1152–63.
Novo D, Heath N, Mitchell L, Caligiuri G, MacFarlane A, Reijmer D, et al. Mutant p53s generate pro-invasive niches by influencing exosome podocalyxin levels. Nat Commun. 2018;9:5069.
Choe SR, Kim YN, Park CG, Cho KH, Cho DY, Lee HY. RCP induces FAK phosphorylation and ovarian cancer cell invasion with inhibition by curcumin. Exp Mol Med. 2018;50:1–10.
Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, Norman JC. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J Cell Biol. 2008;183:143–55.
Hwang MH, Cho KH, Jeong KJ, Park YY, Kim JM, Yu SL, et al. RCP induces Slug expression and cancer cell invasion by stabilizing beta1 integrin. Oncogene. 2017;36:1102–11.
Jacquemet G, Green DM, Bridgewater RE, von Kriegsheim A, Humphries MJ, Norman JC, et al. RCP-driven alpha5beta1 recycling suppresses Rac and promotes RhoA activity via the RacGAP1-IQGAP1 complex. J Cell Biol. 2013;202:917–35.
Morello V, Cabodi S, Sigismund S, Camacho-Leal MP, Repetto D, Volante M, et al. beta1 integrin controls EGFR signaling and tumorigenic properties of lung cancer cells. Oncogene. 2011;30:4087–96.
Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–41.
Lindsay AJ, McCaffrey MW. Rab coupling protein mediated endosomal recycling of N-cadherin influences cell motility. Oncotarget. 2017;8:104717–32.
Lindsay AJ, Hendrick AG, Cantalupo G, Senic-Matuglia F, Goud B, Bucci C, et al. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J Biol Chem. 2002;277:12190–9.
Oduah EI, Grossman SR. Harnessing the vulnerabilities of p53 mutants in lung cancer - Focusing on the proteasome: A new trick for an old foe? Cancer Biol Ther. 2020;21:293–302.
Hafner A, Bulyk ML, Jambhekar A, Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. 2019;20:199–210.
Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–70.
Lozano G. The oncogenic roles of p53 mutants in mouse models. Curr Opin Genet Dev. 2007;17:66–70.
Kim MP, Lozano G. Mutant p53 partners in crime. Cell Death Differ. 2018;25:161–8.
Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–72.
Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–60.
Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, et al. Integrated analysis of TP53 gene and pathway alterations in the cancer genome atlas. Cell Rep. 2019;28:1370–84 e1375.
Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–83.
Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525:206–11.
Escoll M, Gargini R, Cuadrado A, Anton IM, Wandosell F. Mutant p53 oncogenic functions in cancer stem cells are regulated by WIP through YAP/TAZ. Oncogene. 2017;36:3515–27.
Muller PA, Trinidad AG, Timpson P, Morton JP, Zanivan S, van den Berghe PV, et al. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene. 2013;32:1252–65.
Godar S, Ince TA, Bell GW, Feldser D, Donaher JL, Bergh J, et al. Growth-inhibitory and tumor-suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008;134:62–73.
Zhang L, Jin M, Margariti A, Wang G, Luo Z, Zampetaki A, et al. Sp1-dependent Activation of HDAC7 Is Required for Platelet-derived Growth Factor-BB-induced Smooth Muscle Cell Differentiation from Stem Cells. J Biol Chem. 2010;285:38463–72.
Tschaharganeh DF, Xue W, Calvisi DF, Evert M, Michurina TV, Dow LE, et al. p53-dependent nestin regulation links tumor suppression to cellular plasticity in liver cancer. Cell. 2014;158:579–92.
Hwang C-I, Matoso A, Corney DC, Flesken-Nikitin A, Koerner S, Wang W, et al. Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc Natl Acad Sci USA. 2011;108:14240–5.
Mollaoglu G, Guthrie MR, Bohm S, Bragelmann J, Can I, Ballieu PM, et al. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell. 2017;31:270–85.
Kortlever RM, Sodir NM, Wilson CH, Burkhart DL, Pellegrinet L, Brown Swigart L, et al. Myc cooperates with Ras by programming inflammation and immune suppression. Cell. 2017;171:1301–1315 e1314.
Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387:1837–46.
Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl J Med. 2015;372:2018–28.
Gettinger SN, Horn L, Gandhi L, Spigel DR, Antonia SJ, Rizvi NA, et al. Overall survival and long-term safety of nivolumab (Anti-Programmed Death 1 Antibody, BMS-936558, ONO-4538) in patients with previously treated advanced non-small-cell lung cancer. J Clin Oncol. 2015;33:2004–12.
Gundry C, Marco S, Rainero E, Miller B, Dornier E, Mitchell L, et al. Phosphorylation of Rab-coupling protein by LMTK3 controls Rab14-dependent EphA2 trafficking to promote cell:cell repulsion. Nat Commun. 2017;8:14646.
Monteleone E, Orecchia V, Corrieri P, Schiavone D, Avalle L, Moiso E, et al. SP1 and STAT3 functionally synergize to induce the RhoU small GTPase and a subclass of non-canonical WNT responsive genes correlating with poor prognosis in breast cancer. Cancers (Basel). 2019;11:101.
Su HW, Wang SW, Ghishan FK, Kiela PR, Tang MJ. Cell confluency-induced Stat3 activation regulates NHE3 expression by recruiting Sp1 and Sp3 to the proximal NHE3 promoter region during epithelial dome formation. Am J Physiol Cell Physiol. 2009;296:C13–24.
Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: Bring in the family. Genomics. 2005;85:551–6.
Ghosh M, Saha S, Bettke J, Nagar R, Parrales A, Iwakuma T, et al. Mutant p53 suppresses innate immune signaling to promote tumorigenesis. Cancer Cell. 2021;39:494–508 e495.
Muller PAJ, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8.
Zhang J, Sun W, Kong X, Zhang Y, Yang HJ, Ren C, et al. Mutant p53 antagonizes p63/p73-mediated tumor suppression via Notch1. Proc Natl Acad Sci USA. 2019;116:24259–67.
Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98.
Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol. 2011;7:285–95.
Xu S, Tang J, Wang C, Liu J, Fu Y, Luo Y. CXCR7 promotes melanoma tumorigenesis via Src kinase signaling. Cell Death Dis. 2019;10:191.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82103071 to CW) and the National Major Scientific and Technological Special Project for “Significant New Drugs Development” (No. 20181821569 to YL).
Author information
Authors and Affiliations
Contributions
CW and SZ performed all experiments with the assistance from BM and YF. SZ and CW conceived and designed the research. YL supervised the research. C.W and SZ wrote and edited the paper. All authors commented on the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, C., Zhang, S., Ma, B. et al. TP53 mutations upregulate RCP expression via Sp1/3 to drive lung cancer progression. Oncogene 41, 2357–2371 (2022). https://doi.org/10.1038/s41388-022-02260-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02260-7
This article is cited by
-
High FAAP24 expression reveals poor prognosis and an immunosuppressive microenvironment shaping in AML
Cancer Cell International (2023)