Abstract
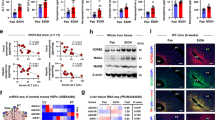
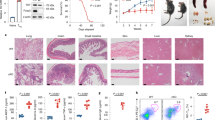
RORγt is a master regulator of Th17 cells. Despite evidence linking RORγt deficiency/inhibition with metastatic thymic T cell lymphomas, the role of RORγt in lymphoma metabolism is unknown. Chronic alcohol consumption plays a causal role in many human cancers. The risk of T cell lymphoma remains unclear in humans with alcohol use disorders (AUD) after chronic RORγt inhibition. Here we demonstrated that alcohol consumption accelerates RORγt deficiency-induced lymphomagenesis. Loss of RORγt signaling in the thymus promotes aerobic glycolysis and glutaminolysis and increases allocation of glutamine carbon into lipids. Importantly, alcohol consumption results in a shift from aerobic glycolysis to glutaminolysis. Both RORγt deficiency- and alcohol-induced metabolic alterations are mediated by c-Myc, as silencing of c-Myc decreases the effects of alcohol consumption and RORγt deficiency on glutaminolysis, biosynthesis, and tumor growth in vivo. The ethanol-mediated c-Myc activation coupled with increased glutaminolysis underscore the critical role of RORγt-Myc signaling and translation in lymphoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33.
He YW, Beers C, Deftos ML, Ojala EW, Forbush KA, Bevan MJ. Down-regulation of the orphan nuclear receptor ROR gamma t is essential for T lymphocyte maturation. J Immunol. 2000;164:5668–74.
Ueda E, Kurebayashi S, Sakaue M, Backlund M, Koller B, Jetten AM. High incidence of T-cell lymphomas in mice deficient in the retinoid-related orphan receptor RORgamma. Cancer Res. 2002;62:901–9.
Guntermann C, Piaia A, Hamel ML, Theil D, Rubic-Schneider T, Del Rio-Espinola A, et al. Retinoic-acid-orphan-receptor-C inhibition suppresses Th17 cells and induces thymic aberrations. JCI Insight. 2017;2:e91127.
Guo Y, MacIsaac KD, Chen Y, Miller RJ, Jain R, Joyce-Shaikh B, et al. Inhibition of RORgammaT skews TCRalpha gene rearrangement and limits T cell repertoire diversity. Cell Rep. 2016;17:3206–18.
Liljevald M, Rehnberg M, Soderberg M, Ramnegard M, Borjesson J, Luciani D, et al. Retinoid-related orphan receptor gamma (RORgamma) adult induced knockout mice develop lymphoblastic lymphoma. Autoimmun Rev. 2016;15:1062–70.
Jun S, Mahesula S, Mathews TP, Martin-Sandoval MS, Zhao Z, Piskounova E, et al. The requirement for pyruvate dehydrogenase in leukemogenesis depends on cell lineage. Cell Metab. 2021;33:1777–92.
Ramstead AG, Wallace JA, Lee SH, Bauer KM, Tang WW, Ekiz HA, et al. Mitochondrial pyruvate carrier 1 promotes peripheral T cell homeostasis through metabolic regulation of thymic development. Cell Rep. 2020;30:2889.
Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20:61–72.
Odera JO, Xiong ZH, Huang CZ, Gu N, Yang WJ, Githang’a J, et al. NRF2/ACSS2 axis mediates the metabolic effect of alcohol drinking on esophageal squamous cell carcinoma. Biochem J. 2020;477:3075–89.
Ma HY, Yamamoto G, Xu J, Liu X, Karin D, Kim JY, et al. IL-17 signaling in steatotic hepatocytes and macrophages promotes hepatocellular carcinoma in alcohol-related liver disease. J Hepatol. 2020;72:946–59.
Zakhari S, Li TK. Determinants of alcohol use and abuse: Impact of quantity and frequency patterns on liver disease. Hepatology. 2007;46:2032–9.
Kishton RJ, Barnes CE, Nichols AG, Cohen S, Gerriets VA, Siska PJ, et al. AMPK is essential to balance glycolysis and mitochondrial metabolism to control T-ALL cell stress and survival. Cell Metab. 2016;23:649–62.
Siler SQ, Neese RA, Christiansen MP, Hellerstein MK. The inhibition of gluconeogenesis following alcohol in humans. Am J Physiol-Endoc M. 1998;275:E897–E907.
Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–21.
Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–7.
Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–U100.
Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Gene Dev. 2006;20:2096–109.
Evangelisti C, Ricci F, Tazzari P, Tabellini G, Battistelli M, Falcieri E, et al. Targeted inhibition of mTORC1 and mTORC2 by active-site mTOR inhibitors has cytotoxic effects in T-cell acute lymphoblastic leukemia. Leukemia. 2011;25:781–91.
Wang Y, Liu Y, Malek SN, Zheng P, Liu Y. Targeting HIF1 alpha eliminates cancer stem cells in hematological malignancies. Cell Stem Cell. 2011;8:399–411.
Chu S, Sun R, Gu X, Chen L, Liu M, Guo H, et al. Inhibition of sphingosine-1-phosphate-induced Th17 cells ameliorates alcoholic steatohepatitis in mice. Hepatology. 2021;73:952–67.
Wang RN, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, et al. The transcription factor myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–82.
Gu Y, Zhang J, Ma X, Kim BW, Wang H, Li J, et al. Stabilization of the c-Myc protein by CAMKIIgamma promotes T cell lymphoma. Cancer Cell. 2017;32:115–28 e117.
Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–79.
Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–34.
DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–50.
Pollizzi KN, Waickman AT, Patel CH, Sun IH, Powell JD. Cellular size as a means of tracking mTOR activity and cell fate of CD4+T cells upon antigen recognition. Plos One. 2015;10:e0121710.
Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–73.
Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–85.
Gao YH, Zhou Z, Ren TY, Kim SJ, He Y, Seo W, et al. Alcohol inhibits T-cell glucose metabolism and hepatitis in ALDH2-deficient mice and humans: roles of acetaldehyde and glucocorticoids. Gut. 2019;68:1311–22.
Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander, et al. Supporting aspartate biosynthesis is an essential function of respiration in proliferating. Cells Cell. 2015;162:552–63.
Mak TW, Grusdat M, Duncan GS, Dostert C, Nonnenmacher Y, Cox M, et al. Glutathione primes T cell metabolism for inflammation. Immunity. 2017;46:675–89.
Choi WM, Kim HH, Kim MH, Cinar R, Yi HS, Eun HS, et al. Glutamate signaling in hepatic stellate cells drives alcoholic steatosis. Cell Metab. 2019;30:877–89 e877.
Laplante M, Sabatini DM. mTOR Signaling in growth control and disease. Cell. 2012;149:274–93.
Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, et al. Small-molecule RORgammat antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 2014;40:477–89.
Scheepstra M, Leysen S, van Almen GC, Miller JR, Piesvaux J, Kutilek V, et al. Identification of an allosteric binding site for RORgammat inhibition. Nat Commun. 2015;6:8833.
Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, et al. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci USA. 2000;97:10132–7.
Yang K, Blanco DB, Chen X, Dash P, Neale G, Rosencrance C, et al. Metabolic signaling directs the reciprocal lineage decisions of alphabeta and gammadelta T cells. Sci Immunol. 2018;3:eaas9818.
Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev. 2012;249:43–58.
Kalaitzidis D, Sykes SM, Wang Z, Punt N, Tang Y, Ragu C, et al. mTOR complex 1 plays critical roles in hematopoiesis and Pten-loss-evoked leukemogenesis. Cell Stem Cell. 2012;11:429–39.
Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, et al. Acetate dependence of tumors. Cell. 2014;159:1591–602.
Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27:57–71.
Nevzorova YA, Cubero FJ, Hu W, Hao F, Haas U, Ramadori P, et al. Enhanced expression of c-myc in hepatocytes promotes initiation and progression of alcoholic liver disease. J Hepatol. 2016;64:628–40.
Shao T, Zhao C, Li F, Gu Z, Liu L, Zhang L, et al. Intestinal HIF-1alpha deletion exacerbates alcoholic liver disease by inducing intestinal dysbiosis and barrier dysfunction. J Hepatol. 2018;69:886–95.
Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–97.
MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–83.
Acknowledgements
This work was supported by grants from the NIH R21AA025724, R21AI128206, R21AI159194 and R01 DK115406 (ZD), and NIH P50AA024337, P20GM113226, (CJM). We thank Dr. J. Ainsworth for editorial assistance.
Author information
Authors and Affiliations
Contributions
ZD and RS designed the research, analyzed and interpreted data, and drafted the manuscript; RS, CL, LC, LH, and HG performed experiments and interpreted data; and WF, XZ, JY, and CJM interpreted the findings and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Sun, R., Lei, C., Chen, L. et al. Alcohol-driven metabolic reprogramming promotes development of RORγt-deficient thymic lymphoma. Oncogene 41, 2287–2302 (2022). https://doi.org/10.1038/s41388-022-02257-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02257-2