Abstract
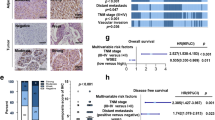
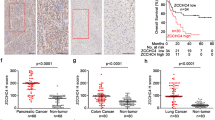
Hepatocellular carcinoma (HCC) is one of the largest causes of cancer-related deaths worldwide owing to the limitation of effective treatment options. The ubiquitin-proteasome system has been rapidly recognized as a frequent target of deregulation leading to cancers. Enhanced DNA damage response (DDR) promotes HCC growth and prevents chemosensitivity, and ubiquitin E3 ligases are key modulators in DDR. Therefore, a better understanding of how E3 ligases regulate cell growth and DNA damage may provide novel insights in understanding the oncogenic mechanism and improving the efficacy of DNA damage therapeutic agents. Here, we performed a high-content RNAi screening targeting 52 DDR-related E3 ligases in HCC and found that ring finger protein 4 (RNF4) was essential for HCC growth. RNF4 was highly expressed in HCC tissues, and the expression levels of RNF4 were associated with poor outcomes. RNF4 silencing significantly suppressed the cell growth, and subsequently induced G2/M arrest and apoptosis of HCC cells in vitro; RNF4 silencing also demonstrated the tumor-suppressive efficacy on HCC in vivo. Moreover, RNF4 silencing increased DNA damage, and rendered HCC cells more sensitive to DNA damage drugs and radiation. We found RNF4 functionally interacts with p62, and mechanistic analyses indicated that RNF4 silencing triggered the nuclear enrichment of p62. Moreover, the p62 nuclear targeting was required for increased DNA damage and growth suppression mediated by RNF4 silencing. Thus, our findings suggest RNF4 is essential for HCC proliferation via preventing nuclear translocation of p62. RNF4 silencing promotes DNA damage and may serve as a novel strategy to suppress cell growth and increase the sensitivity of DNA damage therapeutic agents in HCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Balogh J, Victor D 3rd, Asham EH, Burroughs SG, Boktour M, Saharia A, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41–53.
Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–45.
Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–17.
Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–14.
Wang Y, Zhang N, Zhang L, Li R, Fu W, Ma K, et al. Autophagy regulates chromatin ubiquitination in DNA damage response through elimination of SQSTM1/p62. Mol Cell. 2016;63:34–48.
Sriramachandran AM, Dohmen RJ. SUMO-targeted ubiquitin ligases. Biochim Biophys Acta Mol Cell Res. 2014;1843:75–85.
Vyas R, Kumar R, Clermont F, Helfricht A, Kalev P, Sotiropoulou P, et al. RNF4 is required for DNA double-strand break repair in vivo. Cell Death Differ. 2013;20:490–502.
Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 2012;26:1179–95.
Sánchez-Martín P., Komatsu M. p62/SQSTM1—steering the cell through health and disease. J Cell Sci. 2018;131:jcs222836.
Pankiv S, Lamark T, Bruun JA, Overvatn A, Bjorkoy G, Johansen T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol Chem. 2010;285:5941–53.
Gartner A, Muller S. PML, SUMO, and RNF4: guardians of nuclear protein quality. Mol Cell. 2014;55:1–3.
Brinkmann K, Schell M, Hoppe T, Kashkar H. Regulation of the DNA damage response by ubiquitin conjugation. Front Genet. 2015;6:98.
Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–46.
Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–44.
Goto Y, Koyasu S, Kobayashi M, Harada H. The emerging roles of the ubiquitination/deubiquitination system in tumor radioresistance regarding DNA damage responses, cell cycle regulation, hypoxic responses, and antioxidant properties: insight into the development of novel radiosensitizing strategies. Mutat Res. 2017;803–805:76–81.
Uckelmann M, Sixma TK. Histone ubiquitination in the DNA damage response. DNA Repair. 2017;56:92–101.
Yang D, Zhao Y, Liu J, Sun Y, Jia L. Protective autophagy induced by RBX1/ROC1 knockdown or CRL inactivation via modulating the DEPTOR-MTOR axis. Autophagy. 2012;8:1856–8.
Collins GA, Goldberg AL. The logic of the 26S proteasome. Cell. 2017;169:792–806.
Erpapazoglou Z, Walker O, Haguenauer-Tsapis R. Versatile roles of k63-linked ubiquitin chains in trafficking. Cells. 2014;3:1027–88.
Su Y-T, Gao C, Liu Y, Guo S, Wang A, Wang B, et al. Monoubiquitination of filamin B regulates vascular endothelial growth factor-mediated trafficking of histone deacetylase 7. Mol Cell Biol. 2013;33:1546–60.
Wang G, Gao Y, Li L, Jin G, Cai Z, Chao J-I, et al. K63-linked ubiquitination in kinase activation and cancer. Front Oncol. 2012;2:5–5.
Al-Hakim AK, Zagorska A, Chapman L, Deak M, Peggie M, Alessi DR. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-inked polyubiquitin chains. Biochemical J. 2008;411:249–60.
Sang Y, Yan F, Ren X. The role and mechanism of CRL4 E3 ubiquitin ligase in cancer and its potential therapy implications. Oncotarget. 2015;6:42590–602.
Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18:69–88.
Hao Z, Huang S. E3 ubiquitin ligase Skp2 as an attractive target in cancer therapy. Front Biosci Landmark. 2015;20:474–90.
Jung Y-S, Qian Y, Chen X. Pirh2 RING-finger E3 ubiquitin ligase: Its role in tumorigenesis and cancer therapy. FEBS Lett. 2012;586:1397–402.
Groocock LM, Nie M, Prudden J, Moiani D, Wang T, Cheltsov A, et al. RNF4 interacts with both SUMO and nucleosomes to promote the DNA damage response. Embo Rep. 2014;15:601–8.
Kimberly AF, Xin G, Oliver K, JS O. The Sumo-targeted ubiquitin ligase RNF4 regulates the localization and function of the HTLV-1 oncoprotein Tax. Blood. 2012;119:1173–81.
Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. Febs J. 2015;282:4672–8.
Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–4.
Feng Y, Klionsky DJ. Autophagy regulates DNA repair through SQSTM1/p62. Autophagy. 2017;13:995–6.
Moscat J, Karin M, Diaz-Meco MT. p62 in cancer: signaling adaptor beyond autophagy. Cell. 2016;167:606–9.
Lin X, Li S, Zhao Y, Ma X, Zhang K, He X, et al. Interaction domains of p62: a bridge between p62 and selective autophagy. DNA Cell Biol. 2013;32:220–7.
Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtininduced cell death. J Cell Biol. 2015;171:603–14.
Taniguchi K, Yamachika S, He F, Karin M. p62/SQSTM1-Dr. Jekyll and Mr. Hyde that prevents oxidative stress but promotes liver cancer. FEBS Lett. 2016;590:2375–97.
Bartolini D, Dallaglio K, Torquato P, Piroddi M, Galli F. Nrf2-p62 autophagy pathway and its response to oxidative stress in hepatocellular carcinoma. Transl Res. 2018;193:54–71.
Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–84.
Saito T, Ichimura Y, Taguchi K, Suzuki T, Mizushima T, Takagi K, et al. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun. 2016;7:12030.
Denk H, Stumptner C, Abuja PM, Zatloukal K. Sequestosome 1/p62-related pathways as therapeutic targets in hepatocellular carcinoma. Expert Opin Ther Targets. 2019;23:393–406.
Xiang X, Qin HG, You XM, Wang YY, Qi LN, Ma L, et al. Expression of P62 in hepatocellular carcinoma involving hepatitis B virus infection and aflatoxin B1 exposure. Cancer Med. 2017;6:2357–69.
Zhao XM, Hu WX, Wu ZF, Chen YX, Zeng ZC. Tetrandrine enhances radiosensitization in human hepatocellular carcinoma cell lines. Radiat Res. 2018;190:385–95.
Sun L, Zhang G, Li Z, Lei T, Huang C, Song T, et al. Cellular distribution of tumour suppressor protein p53 and high-risk human papillomavirus (HPV)-18 E6 fusion protein in wild-type p53 cell lines. J Int Med Res. 2008;36:1015–21.
Yi T, Yi Z, Cho SG, Luo J, Pandey MK, Aggarwal BB, et al. Gambogic acid inhibits angiogenesis and prostate tumor growth by suppressing vascular endothelial growth factor receptor 2 signaling. Cancer Res. 2008;68:1843–50.
Su Q, Liu YF, Zhang JF, Zhang SX, Li DF, Yang JJ. Expression of insulin like growth factor II in hepatitis B, cirrhosis and hepatocellular-carcinoma its relationship with hepatitis B virus antigen expression. Hepatology. 1994;20:788–99.
Yang D, Li L, Liu H, Wu L, Luo Z, Li H, et al. Induction of autophagy and senescence by knockdown of ROC1 E3 ubiquitin ligase to suppress the growth of liver cancer cells. Cell Death Differ. 2013;20:235–47.
Fang JH, Zhou HC, Zeng CX, Yang J, Liu YL, Huang XZ, et al. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54:1729–40.
Yang DQ, Zhang Q, Ma YF, Che ZH, Zhang WL, Wu MM, et al. Augmenting the therapeutic efficacy of adenosine against pancreatic cancer by switching the Akt.p21-dependent senescence to apoptosis. EBioMedicine. 2019;47:114–27.
Acknowledgements
This work was supported by National Natural Science Foundation Grant of China [81770579, 81830080, 81861168038, 81970506, 81800477]. The authors thank Ying Zhao for p62 WT, p62 △UBA, p62 △PB1, p62 △NES and p62 △NLS plasmids. The authors thank Dr. Zhaohui Wu and Dr. Mingfang Lv for revising our manuscript. The authors thank Mengxiao Ge for replenishing our experimental data.
Author information
Authors and Affiliations
Contributions
BL and YP contributed to acquisition, analysis and interpretation of data, and original draft writing. DH, PC, ML, and ZY contributed to acquisition of data. JZ contributed to analysis and interpretation of data. YC contributed to article revision. DY and JL contributed to study design and article revision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lv, B., Pan, Y., Hou, D. et al. RNF4 silencing induces cell growth arrest and DNA damage by promoting nuclear targeting of p62 in hepatocellular carcinoma. Oncogene 41, 2275–2286 (2022). https://doi.org/10.1038/s41388-022-02247-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02247-4