Abstract
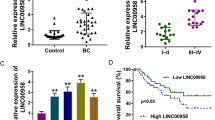
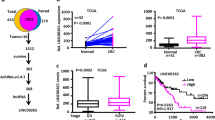
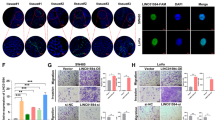
Insulin-like growth factor-2 mRNA-binding protein 2 (IGF2BP2, also known as IMP2), a novel class III N6-methyladenosine (m6A) reader, has recently gained attention due to its critical functions in recognizing and stabilizing m6A modified oncogenic transcripts. However, whether and how long non-coding RNAs (lncRNAs) facilitate IMP2’s role as m6A “reader” remains elusive, particularly in colorectal cancer (CRC). Here, we demonstrated that oncogenic LINC021 specifically bound with the m6A “reader” IMP2 protein and enhanced the mRNA stability of MSX1 and JARID2 in an m6A regulatory manner during CRC tumorigenesis and pathogenesis. Specifically, a remarkable upregulation of LINC021 was confirmed in CRC cell lines and clinical tissues (n = 130). High level of LINC021acted as an independent prognostic predictor for CRC clinical outcomes. Functional assays demonstrated that LINC021 exerted its functions as an oncogene to aggravate CRC malignant phenotypes including enhanced cell proliferation, colony formation, migration capabilities, and reduced cell apoptosis. Mechanistically, LINC021 directly recognized IMP2 protein, the latter enhanced the mRNA stability of transcripts such as MSX1 and JARID2 by recognizing their m6A-modified element RGGAC. Thus, these findings uncovered an essential LINC021/IMP2/MSX1 and JARID2 signaling axis in CRC tumorigenesis, which provided profound insights into our understanding of m6A modification regulated by lncRNA in CRC initiation and progression and shed light on the targeting of this axis for CRC treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The authors declare that all the data supporting the findings in this study are available in this study and its Supplementary materials, or are available from the corresponding author through reasonable request. The Raw data were download from the Gene Expression Omnibus (GEO) website, show as follows:
References
Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell. 2020;37:270–88.
Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12:121.
Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol Cancer. 2020;19:88.
He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176.
Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573–6.
Yi YC, Chen XY, Zhang J, Zhu JS. Novel insights into the interplay between m(6)A modification and noncoding RNAs in cancer. Mol Cancer. 2020;19:121.
Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–17.
Roignant JY, Soller M. m(6)A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 2017;33:380–90.
Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74:640–50.
Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–24.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95.
Chatterji P, Rustgi AK. RNA binding proteins in intestinal epithelial biology and colorectal cancer. Trends Mol Med. 2018;24:490–506.
Degrauwe N, Suva ML, Janiszewska M, Riggi N, Stamenkovic I. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 2016;30:2459–74.
Muller S, Glass M, Singh AK, Haase J, Bley N, Fuchs T, et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019;47:375–90.
Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–205.
Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112.
Qu F, Tsegay PS, Liu Y. N(6)-methyladenosine, DNA repair, and genome stability. Front Mol Biosci. 2021;8:645823.
Wang Y, Wang Y, Luo W, Song X, Huang L, Xiao J, et al. Roles of long non-coding RNAs and emerging RNA-binding proteins in innate antiviral responses. Theranostics. 2020;10:9407–24.
Liu H, Xu Y, Yao B, Sui T, Lai L, Li Z. A novel N6-methyladenosine (m6A)-dependent fate decision for the lncRNA THOR. Cell Death Dis. 2020;11:613.
Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27:1782–94.
Ban Y, Tan P, Cai J, Li J, Hu M, Zhou Y, et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol Oncol. 2020;14:1282–96.
Hunten S, Kaller M, Drepper F, Oeljeklaus S, Bonfert T, Erhard F, et al. p53-regulated networks of protein, mRNA, miRNA, and lncRNA expression revealed by integrated pulsed stable isotope labeling with amino acids in cell culture (pSILAC) and next generation sequencing (NGS) analyses. Mol Cell Proteom. 2015;14:2609–29.
Zhou D, Tian F, Tian X, Sun L, Huang X, Zhao F, et al. Diagnostic evaluation of a deep learning model for optical diagnosis of colorectal cancer. Nat Commun. 2020;11:2961.
Francescangeli F, De Angelis ML, Zeuner A. Dietary factors in the control of gut homeostasis, intestinal stem cells, and colorectal cancer. Nutrients. 2019;11:2936
Marmol I, Sanchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18:197
Meyer J, Orci LA, Combescure C, Balaphas A, Morel P, Buchs NC, et al. Risk of colorectal cancer in patients with acute diverticulitis: a systematic review and meta-analysis of observational studies. Clin Gastroenterol Hepatol. 2019;17:1448–56.e1417.
Zhao Y, Chen Y, Jin M, Wang J. The crosstalk between m(6)A RNA methylation and other epigenetic regulators: a novel perspective in epigenetic remodeling. Theranostics. 2021;11:4549–66.
Dai F, Wu Y, Lu Y, An C, Zheng X, Dai L, et al. Crosstalk between RNA m(6)A modification and non-coding RNA contributes to cancer growth and progression. Mol Ther Nucleic Acids. 2020;22:62–71.
Khan RIN, Malla WA. m(6)A modification of RNA and its role in cancer, with a special focus on lung cancer. Genomics. 2021;113:2860–9.
Zhang J, Hu K, Yang YQ, Wang Y, Zheng YF, Jin Y, et al. LIN28B-AS1-IGF2BP1 binding promotes hepatocellular carcinoma cell progression. Cell Death Dis. 2020;11:741.
Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX, et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18:174.
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18:143.
Zhang Y, Geng X, Li Q, Xu J, Tan Y, Xiao M, et al. m6A modification in RNA: biogenesis, functions and roles in gliomas. J Exp Clin Cancer Res. 2020;39:192.
Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D, et al. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol Cancer. 2020;19:104.
Zhao W, Qi X, Liu L, Ma S, Liu J, Wu J. Epigenetic regulation of m(6)A modifications in human cancer. Mol Ther Nucleic Acids. 2020;19:405–12.
Zaccara S, Jaffrey SR. A unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell. 2020;181:1582–95.e1518.
Tong J, Flavell RA, Li HB. RNA m(6)A modification and its function in diseases. Front Med. 2018;12:481–9.
Lu S, Han L, Hu X, Sun T, Xu D, Li Y, et al. N6-methyladenosine reader IMP2 stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect: implication in colorectal cancer. J Hematol Oncol. 2021;14:188.
Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cance. Int J Mol Sci. 2019;20:5758.
Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17:106–16.
Zhou B, Yang H, Yang C, Bao YL, Yang SM, Liu J, et al. Translation of noncoding RNAs and cancer. Cancer Lett. 2021;497:89–99.
Hou P, Meng S, Li M, Lin T, Chu S, Li Z, et al. LINC00460/DHX9/IGF2BP2 complex promotes colorectal cancer proliferation and metastasis by mediating HMGA1 mRNA stability depending on m6A modification. J Exp Clin Cancer Res. 2021;40:52.
Zhu P, He F, Hou Y, Tu G, Li Q, Jin T, et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene. 2021;40:1609–27.
Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X, et al. m(6)A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19:72.
Heppt MV, Wang JX, Hristova DM, Wei Z, Li L, Evans B, et al. MSX1-induced neural crest-like reprogramming promotes melanoma progression. J Invest Dermatol. 2018;138:141–9.
Adhikari A, Mainali P, Davie JK. JARID2 and the PRC2 complex regulate the cell cycle in skeletal muscle. J Biol Chem. 2019;294:19451–64.
Acknowledgements
We gratefully appreciate the efforts and contributions of doctors, nurses, and technical staff at the First Hospital of China Medical University, Cancer Hospital of China Medical University.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81872905, 82073884), Science and Technology Innovative Foundation for Young and Middle-aged Scientists of Shenyang City (RC200382), Shenyang High Level Talent Innovation and Entrepreneurship Team (2019-SYRCCY-B-01), and Major Special S&T Projects in Liaoning Province [2019JH1/10300005].
Author information
Authors and Affiliations
Contributions
HW, MW, and XD conceived and designed the project. HW, XD, QC, TS, XH, HG, ML, ZG, WY, LZ, KL performed experiments and/or data acquisition and analyses; HW, XD, QC, TS, XH, HG, ML, KL contributed technical/reagents materials, analytic tools and/or grant support; HW, MW, KL, and XD prepared, wrote, and/or revision the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of China Medical University. The animal experiments (CMU2020184) performed in this study were approved by the Institutional Animal Care and Use Committee of China Medical University.
Consent for publication
All of the authors have written informed consent.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wu, H., Ding, X., Hu, X. et al. LINC01021 maintains tumorigenicity by enhancing N6-methyladenosine reader IMP2 dependent stabilization of MSX1 and JARID2: implication in colorectal cancer. Oncogene 41, 1959–1973 (2022). https://doi.org/10.1038/s41388-022-02189-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02189-x
This article is cited by
-
The m6A modification mediated-lncRNA POU6F2-AS1 reprograms fatty acid metabolism and facilitates the growth of colorectal cancer via upregulation of FASN
Molecular Cancer (2024)
-
FBW7/GSK3β mediated degradation of IGF2BP2 inhibits IGF2BP2-SLC7A5 positive feedback loop and radioresistance in lung cancer
Journal of Experimental & Clinical Cancer Research (2024)
-
Oncofetal protein IGF2BPs in human cancer: functions, mechanisms and therapeutic potential
Biomarker Research (2023)
-
JMJD family proteins in cancer and inflammation
Signal Transduction and Targeted Therapy (2022)