Abstract
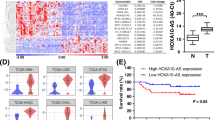
MicroRNAs (miRNAs) may modulate more than 60% of human coding genes and act as negative regulators, whereas long noncoding RNAs (lncRNAs) regulate gene expression on multiple levels by interacting with chromatin, functional proteins, and RNAs such as mRNAs and microRNAs. However, the crosstalk between HOTTIP lncRNA and miRNAs in leukemogenesis remains elusive. Using combined integrated analyses of global miRNA expression profiling and state-of-the-art genomic analyses of chromatin such as ChIRP-seq (HOTTIP binding in genomewide), ChIP-seq, and ATAC-seq, we found that some miRNA genes are directly controlled by HOTTIP. Specifically, the HOX cluster miRNAs (miR-196a, miR-196b, miR-10a, and miR-10b), located cis and trans, were most dramatically regulated and significantly decreased in HOTTIP−/− AML cells. HOTTIP bound to the miR-196b promoter and HOTTIP deletion reduced chromatin accessibility and enrichment of active histone modifications at HOX cluster-associated miRNAs in AML cells, whereas reactivation of HOTTIP restored miR gene expression and chromatin accessibility in the CTCF-boundary-attenuated AML cells. Inactivation of HOTTIP or miR-196b promotes apoptosis by altering the chromatin signature at the FAS promoter and increasing FAS expression. Transplantation of miR-196b knockdown MOLM13 cells in NSG mice increased overall survival of mice compared to wild-type cells transplanted into mice. Thus, HOTTIP remodels the chromatin architecture around miRNAs to promote their transcription and consequently represses tumor suppressors and promotes leukemogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
He RZ, Luo DX, Mo YY. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019;6:6–15.
Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5:e944014.
Lambert M, Alioui M, Jambon S, Depauw S, Van Seuningen I, David-Cordonnier MH. Direct and indirect targeting of HOXA9 transcription factor in acute myeloid leukemia. Cancers (Basel). 2019;11:837.
Collins CT, Hess JL. Deregulation of the HOXA9/MEIS1 axis in acute leukemia. Curr Opin Hematol. 2016;23:354–61.
De Kumar B, Krumlauf R. HOXs and lincRNAs: two sides of the same coin. Sci Adv. 2016;2:e1501402.
Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription. Sci Adv. 2017;3:eaao2110.
Grossi E, Raimondi I, Goni E, Gonzalez J, Marchese FP, Chapaprieta V, et al. A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat Commun. 2020;11:936.
Singh AP, Archer TK. Analysis of the SWI/SNF chromatin-remodeling complex during early heart development and BAF250a repression cardiac gene transcription during P19 cell differentiation. Nucleic Acids Res. 2014;42:2958–75.
Singh AP, Foley JF, Rubino M, Boyle MC, Tandon A, Shah R, et al. Brg1 enables rapid growth of the early embryo by suppressing genes that regulate apoptosis and cell growth arrest. Mol Cell Biol. 2016;36:1990–2010.
Singh AP, Foley J, Tandon A, Phadke D, Karimi Kinyamu H, Archer TK. A role for BRG1 in the regulation of genes required for development of the lymphatic system. Oncotarget. 2017;8:54925–38.
Basta JM, Singh A, Robbins L, Stout L, Pherson M, Rauchman M. The core SWI/SNF catalytic subunit Brg1 regulates nephron progenitor cell proliferation and differentiation. Dev Biol. 2020;464:176–87.
Luo H, Zhu G, Xu J, Lai Q, Yan B, Guo Y, et al. HOTTIP lncRNA promotes hematopoietic stem cell self-renewal leading to AML-like disease in mice. Cancer Cell. 2019;36:645–59 e648.
Singh AP, Hung YH, Shanahan MT, Kanke M, Bonfini A, Dame MK, et al. Enteroendocrine progenitor cell-enriched miR-7 regulates intestinal epithelial proliferation in an Xiap-dependent manner. Cell Mol Gastroenterol Hepatol. 2020;9:447–64.
Fernandes JCR, Acuna SM, Aoki JI, Floeter-Winter LM, Muxel SM. Long non-coding RNAs in the regulation of gene expression: physiology and disease. Noncoding RNA. 2019;5:17.
Furio-Tari P, Tarazona S, Gabaldon T, Enright AJ, Conesa A. spongeScan: a web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res. 2016;44:W176–180.
Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15:177–86.
Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–33.
Yendamuri S, Calin GA. The role of microRNA in human leukemia: a review. Leukemia. 2009;23:1257–63.
Wallace JA, O’Connell RM. MicroRNAs and acute myeloid leukemia: therapeutic implications and emerging concepts. Blood. 2017;130:1290–301.
Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci USA. 2006;103:5078–83.
Tan Y, Zhang B, Wu T, Skogerbo G, Zhu X, Guo X, et al. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol. 2009;10:12.
Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI, et al. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137:2136–45. e2131-2137
Stadthagen G, Tehler D, Hoyland-Kroghsbo NM, Wen J, Krogh A, Jensen KT, et al. Loss of miR-10a activates lpo and collaborates with activated Wnt signaling in inducing intestinal neoplasia in female mice. PLoS Genet. 2013;9:e1003913.
Neijts R, Amin S, van Rooijen C, Tan S, Creyghton MP, de Laat W, et al. Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos. Genes Dev. 2016;30:1937–42.
Rodrigues AR, Yakushiji-Kaminatsui N, Atsuta Y, Andrey G, Schorderet P, Duboule D, et al. Integration of Shh and Fgf signaling in controlling Hox gene expression in cultured limb cells. Proc Natl Acad Sci USA. 2017;114:3139–44.
Denans N, Iimura T, Pourquie O. Hox genes control vertebrate body elongation by collinear Wnt repression. Elife. 2015;4:e04379.
Takacs-Vellai K, Vellai T, Chen EB, Zhang Y, Guerry F, Stern MJ, et al. Transcriptional control of Notch signaling by a HOX and a PBX/EXD protein during vulval development in C. elegans. Dev Biol. 2007;302:661–9.
Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012;3:688.
Mo JS, Alam KJ, Kang IH, Park WC, Seo GS, Choi SC, et al. MicroRNA 196B regulates FAS-mediated apoptosis in colorectal cancer cells. Oncotarget. 2015;6:2843–55.
Luo H, Wang F, Zha J, Li H, Yan B, Du Q, et al. CTCF boundary remodels chromatin domain and drives aberrant HOX gene transcription in acute myeloid leukemia. Blood. 2018;132:837–48.
Qiu Y, Xu M, Huang S. Long noncoding RNAs: emerging regulators of normal and malignant hematopoiesis. Blood. 2021;blood.2021011992.
Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 2018;75:467–84.
Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4.
de Boer J, Walf-Vorderwulbecke V, Williams O. In focus: MLL-rearranged leukemia. Leukemia. 2013;27:1224–8.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302.
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501.
Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–93.
Schep AN, Wu BJ, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods. 2017;14:975–+.
Acknowledgements
We are grateful to the Penn State College of Medicine Genome Science Facility for illumine sequencing core and the Flow Cytometry & Cell Sorting Core. We are grateful for helpful discussions from Dr. Yi Qiu (Penn State College of Medicine). We thank Dr. Sachin Singh (Center for Cellular and Molecular Biology, India) for helping the data analysis from the TCGA dataset. This work was supported by the grants from National Institutes of Health (S.H., R01DK110108, R01CA204044, and R01HL141950).
Author information
Authors and Affiliations
Contributions
Conceptualization, study design and direction, performed overall experiments, data analysis, result interpretation, writing original draft and editing of the manuscript, and project administration: APS, HL, MM, ME, KH, and SH. Data curation: HL, MM, ME KH, and AS. Manuscript review, editing, and data visualization: HL, ME, and SH. Project administration: SH. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Singh, A.P., Luo, H., Matur, M. et al. A coordinated function of lncRNA HOTTIP and miRNA-196b underpinning leukemogenesis by targeting FAS signaling. Oncogene 41, 718–731 (2022). https://doi.org/10.1038/s41388-021-02127-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02127-3
This article is cited by
-
Exploration and bioinformatic prediction for profile of mRNA bound to circular RNA BTBD7_hsa_circ_0000563 in coronary artery disease
BMC Cardiovascular Disorders (2024)
-
Association of Genetic Polymorphisms in Long Noncoding RNA HOTTIP with Risk of Idiopathic Recurrent Spontaneous Abortion
Biochemical Genetics (2023)