Abstract
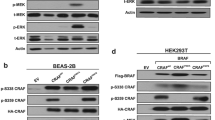
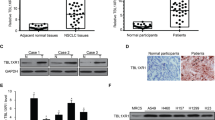
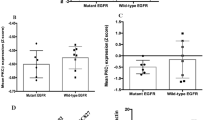
We previously found the SLC3A2-NRG1 (S-N) fusion gene in a lung adenocarcinoma specimen without known driver mutations and validated this in 59 invasive mucinous adenocarcinoma (IMA) samples. Interestingly, KRAS mutation coexisted (62.5%) in 10 out of 16 NRG1 fusions. In this study, we examined the role of mutant KRAS in regulating the S-N fusion protein in KRAS mutant (H358) and wild-type (Calu-3) cells. KRAS mutation-mediated increase in MEK1/2 and ERK1/2 activity enhanced disintegrin and metalloproteinase (ADAM)17 activity, which increased the shedding of NRG1 from the S-N fusion protein. The cleavage of NRG1 also increased the phosphorylation of ERBB2-ERBB3 heterocomplex receptors and their downstream signalling pathways, including PI3K/Akt/mTOR, even under activated KRAS mutation signalling. The concurrence of S-N fusion and KRAS mutation synergistically increased cell proliferation, colony formation, tumour growth, and the cells’ resistance to EGFR kinase inhibitors more than KRAS mutation alone. Targeted inhibition of MEK1/2, and ADAM17 significantly induced apoptosis singly and when combined with each mutation singly or with chemotherapy in both the concurrent KRAS mutant and S-N fusion xenograft and lung orthotopic models. Taken together, this is the first study to report that KRAS mutation increased NRG1 cleavage from the S-N fusion protein through ADAM17, thereby enhancing the Ras/Raf/MEK/ERK and ERBB/PI3K/Akt/mTOR pathways. Moreover, the coexistence of KRAS mutant and S-N fusion in lung tumours renders them vulnerable to MEK1/2 and/or ADAM17 inhibitors, at least in part, due to their dependency on the strong positive loop between KRAS mutation and S-N fusion.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Wong MC, Lao XQ, Ho K-F, Goggins WB, Shelly L. Incidence and mortality of lung cancer: Global trends and association with socioeconomic status. Sci Rep. 2017;7:1–9.
Group NM-aC. Preoperative chemotherapy for non-small-cell lung cancer: A systematic review and meta-analysis of individual participant data. Lancet. 2014;383:1561–71.
Bos JL. Ras oncogenes in human cancer: A review. Cancer Res. 1989;49:4682–9.
Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J cell Sci. 2005;118:843–6.
Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–81.
DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064.
Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–51.
Saad MI, Alhayyani S, McLeod L, Yu L, Alanazi M, Deswaerte V, et al. ADAM 17 selectively activates the IL‐6 trans‐signaling/ERK MAPK axis in KRAS‐addicted lung cancer. EMBO Mol Med. 2019;11:e9976.
Zunke F, Rose-John S. The shedding protease ADAM17: Physiology and pathophysiology. Biochimica et Biophysica Acta (BBA)-Mol Cell Res. 2017;1864:2059–70.
Saad MI, Rose-John S, Jenkins BJ. ADAM17: An emerging therapeutic target for lung cancer. Cancers 2019;11:1218.
Van Schaeybroeck S, Kyula JN, Fenton A, Fenning CS, Sasazuki T, Shirasawa S, et al. Oncogenic Kras promotes chemotherapy-induced growth factor shedding via ADAM17. Cancer Res. 2011;71:1071–80.
Ardito CM, Grüner BM, Takeuchi KK, Lubeseder-Martellato C, Teichmann N, Mazur PK, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer cell. 2012;22:304–17.
Shin DH, Lee D, Hong DW, Hong SH, Hwang J-A, Lee BI, et al. Oncogenic function and clinical implications of SLC3A2-NRG1 fusion in invasive mucinous adenocarcinoma of the lung. Oncotarget. 2016;7:69450.
Shin DH, Jo JY, Han J-Y. Dual targeting of ERBB2/ERBB3 for the treatment of SLC3A2-NRG1–mediated lung cancer. Mol Cancer Therapeutics. 2018;17:2024–33.
Travis WD, Brambilla E, Riely GJ. New pathologic classification of lung cancer: Relevance for clinical practice and clinical trials. J Clin Oncol. 2013;31:992–1001.
Jonna S, Feldman RA, Swensen J, Gatalica Z, Korn WM, Borghaei H, et al. Detection of NRG1 gene fusions in solid tumors. Clin Cancer Res. 2019;25:4966–72.
Chang JC, Offin M, Falcon CJ, Brown DN, Loomis B, Meng F. et al. Comprehensive molecular and clinicopathologic analysis of 200 pulmonary invasive mucinous adenocarcinomas identifies distinct characteristics of molecular subtypes. Clin Cancer Res. 2021;27:4066–76.
Unni AM, Lockwood WW, Zejnullahu K, Lee-Lin S-Q, Varmus H. Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. Elife. 2015;4:e06907.
Ambrogio C, Barbacid M, Santamaría D. In vivo oncogenic conflict triggered by co-existing KRAS and EGFR activating mutations in lung adenocarcinoma. Oncogene 2017;36:2309–18.
Itahana K, Campisi J, Dimri GP. Methods to detect biomarkers of cellular senescence:the senescence-associated beta-galactosidase assay. Method Mol Biol. 2007;371:21–31.
Freed DM, Bessman NJ, Kiyatkin A, Salazar-Cavazos E, Byrne PO, Moore JO, et al. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell 2017;171:683–95. e18
Chen J, Zeng F, Forrester SJ, Eguchi S, Zhang M-Z, Harris RC. Expression and function of the epidermal growth factor receptor in physiology and disease. Physiological Rev. 2016;96:1025–69.
Kim TM, Song A, Kim D-W, Kim S, Ahn Y-O, Keam B, et al. Mechanisms of acquired resistance to AZD9291: A mutation-selective, irreversible EGFR inhibitor. J Thorac Oncol. 2015;10:1736–44.
Ma S, Zhang L, Ren Y, Dai W, Chen T, Luo L, et al. Epiregulin confers EGFR-TKI resistance via EGFR/ErbB2 heterodimer in non-small cell lung cancer. Oncogene 2021;40:2596–609.
Van Schaeybroeck S, Kyula JN, Fenton A, Fenning CS, Sasazuki T, Shirasawa S, et al. Oncogenic Kras promotes chemotherapy-induced growth factor shedding via ADAM17. Cancer Res. 2011;71:1071–80.
Kim BM. The role of saikosaponins in therapeutic strategies for age-related diseases. Oxidative Med. Cell. Longev. 2018;2018:8275256.
Xu P, Derynck R. Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol cell. 2010;37:551–66.
Kwan JC, Eksioglu EA, Liu C, Paul VJ, Luesch H. Grassystatins A− C from marine cyanobacteria, potent cathepsin E inhibitors that reduce antigen presentation. J medicinal Chem. 2009;52:5732–47.
Janssens E, Gaublomme D, De Groef L, Darras VM, Arckens L, Delorme N, et al. Matrix metalloproteinase 14 in the zebrafish: An eye on retinal and retinotectal development. PloS one. 2013;8:e52915.
Zheng Z-Y, Anurag M, Lei JT, Cao J, Singh P, Peng J, et al. Neurofibromin is an estrogen receptor-α transcriptional co-repressor in breast cancer. Cancer Cell. 2020;37:387–402. e7
Hames ML, Chen H, Iams W, Aston J, Lovly CM, Horn L. Correlation between KRAS mutation status and response to chemotherapy in patients with advanced non-small cell lung cancer☆. Lung Cancer. 2016;92:29–34.
Greulich H. The genomics of lung adenocarcinoma: Opportunities for targeted therapies. Genes Cancer. 2010;1:1200–10.
Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. Jama. 2014;311:1998–2006.
Cisowski J, Sayin V, Liu M, Karlsson C, Bergo M. Oncogene-induced senescence underlies the mutual exclusive nature of oncogenic KRAS and BRAF. Oncogene. 2016;35:1328–33.
You JS, Jones PA. Cancer genetics and epigenetics: Two sides of the same coin? Cancer cell. 2012;22:9–20.
Torti D, Trusolino L. Oncogene addiction as a foundational rationale for targeted anti‐cancer therapy: Promises and perils. EMBO Mol Med. 2011;3:623–36.
Kamb A. Consequences of nonadaptive alterations in cancer. Mol Biol cell. 2003;14:2201–5.
Sharma SV, Settleman J. Exploiting the balance between life and death: Targeted cancer therapy and “oncogenic shock”. Biochemical Pharmacol. 2010;80:666–73.
Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–98.
Karreth FA, Tuveson DA. Modelling oncogenic Ras/Raf signalling in the mouse. Curr Opin Genet Dev. 2009;19:4–11.
Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–67.
Ying H, Dey P, Yao W, Kimmelman AC, Draetta GF, Maitra A, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30:355–85.
Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, et al. Essential role for oncogenic Ras in tumour maintenance. Nature 1999;400:468–72.
Gschwind A, Hart S, Fischer OM, Ullrich A. TACE cleavage of proamphiregulin regulates GPCR‐induced proliferation and motility of cancer cells. EMBO J. 2003;22:2411–21.
Ni S-S, Zhang J, Zhao W-L, Dong X-C, Wang J-L. ADAM17 is overexpressed in non-small cell lung cancer and its expression correlates with poor patient survival. Tumor Biol. 2013;34:1813–8.
Zhou B-BS, Peyton M, He B, Liu C, Girard L, Caudler E, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50.
Baumgart A, Seidl S, Vlachou P, Michel L, Mitova N, Schatz N, et al. ADAM17 regulates epidermal growth factor receptor expression through the activation of Notch1 in non–small cell lung cancer. Cancer Res. 2010;70:5368–78.
Sharma A, Bender S, Zimmermann M, Riesterer O, Broggini-Tenzer A, Pruschy MN. Secretome signature identifies ADAM17 as novel target for radiosensitization of non–small cell lung cancer. Clin Cancer Res. 2016;22:4428–39.
Fridman JS, Caulder E, Hansbury M, Liu X, Yang G, Wang Q, et al. Selective inhibition of ADAM metalloproteases as a novel approach for modulating ErbB pathways in cancer. Clin Cancer Res. 2007;13:1892–902.
Acknowledgements
This work was supported by grants (NCC-2010070 and 2110340) funded by the National Cancer Centre, and NRF-2021R1F1A1062721 funded by the National Research Fund.
Author information
Authors and Affiliations
Contributions
Conception and design: DHS and JYH. Acquisition of data: DHS, YGB, SHK, and MYC. Analysis and interpretation of data: DHS, BKC, CYH, SSK. Writing and revision of the paper: DHS.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Shin, D.H., Kim, S.H., Choi, M. et al. Oncogenic KRAS promotes growth of lung cancer cells expressing SLC3A2-NRG1 fusion via ADAM17-mediated shedding of NRG1. Oncogene 41, 280–292 (2022). https://doi.org/10.1038/s41388-021-02097-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02097-6
This article is cited by
-
Examining the contribution of Notch signaling to lung disease development
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)