Abstract
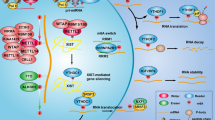
Gastric cancer (GC) has the fifth highest incidence globally, but its molecular mechanisms are not well understood. Here, we report that coactivator-associated arginine methyltransferase 1 (CARM1) is specifically highly expressed in gastric cancer and that its overexpression correlates with poor prognosis in patients with gastric cancer. Nucleoporin 54 (Nup54) was identified as a CARM1-interacting protein that promoted CARM1 nuclear importation. In the nucleus, CARM1 cooperates with transcriptional factor EB (TFEB) to activate Notch2 transcription by inducing H3R17me2 of the Notch2 promoter but not H3R26me2. Additionally, the Notch2 intracellular domain (N2ICD) was identified as a CARM1 substrate. Methylation of N2ICD at R1786, R1838, and R2047 by CARM1 enhanced the binding between N2ICD and mastermind-like protein 1 (MAML1) and increased gastric cancer cell proliferation in vitro and tumor formation in vivo. Our findings reveal a molecular mechanism linking CARM1-mediated transcriptional activation of the Notch2 signaling pathway to Notch2 methylation in gastric cancer progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–72.
Larsen SC, Sylvestersen KB, Mund A, Lyon D, Mullari M, Madsen MV, et al. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci Signal. 2016;9:rs9.
Yadav N, Cheng D, Richard S, Morel M, Iyer VR, Aldaz CM, et al. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 2008;9:193–8.
Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol Cell. 2009;33:1–13.
Blanc RS, Richard S. Arginine Methylation: The Coming of age. Mol Cell. 2017;65:8–24.
Wang L, Zeng H, Wang Q, Zhao Z, Boyer TG, Bian X, et al. MED12 methylation by CARM1 sensitizes human breast cancer cells to chemotherapy drugs. Sci Adv. 2015;1:e1500463.
Kawabe Y, Wang YX, McKinnell IW, Bedford MT, Rudnicki MA. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell. 2012;11:333–45.
Wu Q, Bruce AW, Jedrusik A, Ellis PD, Andrews RM, Langford CF, et al. CARM1 is required in embryonic stem cells to maintain pluripotency and resist differentiation. Stem Cells. 2009;27:2637–45.
Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–8.
Kim YR, Lee BK, Park RY, Nguyen NT, Bae JA, Kwon DD, et al. Differential CARM1 expression in prostate and colorectal cancers. BMC Cancer. 2010;10:197.
Gao WW, Xiao RQ, Zhang WJ, Hu YR, Peng BL, Li WJ, et al. JMJD6 licenses ERalpha-dependent enhancer and coding gene activation by modulating the recruitment of the CARM1/MED12 Co-activator Complex. Mol Cell. 2018;70:340–57 e348.
El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, et al. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc Natl Acad Sci USA. 2006;103:13351–6.
Zhong XY, Yuan XM, Xu YY, Yin M, Yan WW, Zou SW, et al. CARM1 Methylates GAPDH to regulate glucose metabolism and is suppressed in liver cancer. Cell Rep. 2018;24:3207–23.
Wang YP, Zhou W, Wang J, Huang X, Zuo Y, Wang TS, et al. Arginine Methylation of MDH1 by CARM1 inhibits Glutamine Metabolism and suppresses pancreatic cancer. Mol Cell. 2016;64:673–87.
Liu F, Ma F, Wang Y, Hao L, Zeng H, Jia C, et al. PKM2 methylation by CARM1 activates aerobic glycolysis to promote tumorigenesis. Nat Cell Biol. 2017;19:1358–70.
Wang L, Zhao Z, Meyer MB, Saha S, Yu M, Guo A, et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell. 2014;25:21–36.
Cheng D, Vemulapalli V, Lu Y, Shen J, Aoyagi S, Fry CJ, et al. CARM1 methylates MED12 to regulate its RNA-binding ability. Life Sci Alliance. 2018;1:e201800117.
Shishkova E, Zeng H, Liu F, Kwiecien NW, Hebert AS, Coon JJ, et al. Global mapping of CARM1 substrates defines enzyme specificity and substrate recognition. Nat Commun. 2017;8:15571.
Liu J, Feng J, Li L, Lin L, Ji J, Lin C, et al. Arginine methylation-dependent LSD1 stability promotes invasion and metastasis of breast cancer. EMBO Rep. 2020;21:e48597.
Karakashev S, Zhu H, Wu S, Yokoyama Y, Bitler BG, Park PH, et al. CARM1-expressing ovarian cancer depends on the histone methyltransferase EZH2 activity. Nat Commun. 2018;9:631.
Hazawa M, Lin DC, Kobayashi A, Jiang YY, Xu L, Dewi FRP, et al. ROCK-dependent phosphorylation of NUP62 regulates p63 nuclear transport and squamous cell carcinoma proliferation. EMBO Rep. 2018;19:73–88.
Borlido J, D’Angelo MA. Nup62-mediated nuclear import of p63 in squamous cell carcinoma. EMBO Rep. 2018;19:3–4.
Chug H, Trakhanov S, Hulsmann BB, Pleiner T, Gorlich D. Crystal structure of the metazoan Nup62*Nup58*Nup54 nucleoporin complex. Science. 2015;350:106–10.
Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–7.
Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50.
Shin HJ, Kim H, Oh S, Lee JG, Kee M, Ko HJ, et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534:553–7.
Haines N, Irvine KD. Glycosylation regulates notch signalling. Nat Rev Mol Cell Biol. 2003;4:786–97.
Hunter GL, Giniger E. Phosphorylation and proteolytic cleavage of notch in canonical and noncanonical notch signaling. Adv Exp Med Biol. 2020;1227:51–68.
Lim R, Sugino T, Nolte H, Andrade J, Zimmermann B, Shi C, et al. Deubiquitinase USP10 regulates Notch signaling in the endothelium. Science. 2019;364:188–93.
Jeong GY, Park MK, Choi HJ, An HW, Park YU, Choi HJ, et al. NSD3-induced Methylation of H3K36 Activates NOTCH signaling to drive breast tumor initiation and metastatic progression. Cancer Res. 2021;81:77–90.
Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137:216–33.
Andersen P, Uosaki H, Shenje LT, Kwon C. Non-canonical notch signaling: Emerging role and mechanism. Trends Cell Biol. 2012;22:257–65.
Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–9.
Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, et al. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr Biol: CB. 1995;5:1416–23.
Wang L, Zhao Z, Meyer MB, Saha S, Yu M, Guo A, et al. CARM1 Methylates Chromatin Remodeling Factor BAF155 to Enhance Tumor Progression and Metastasis. Cancer Cell. 2016;30:179–80.
Thandapani P, O’Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol Cell. 2013;50:613–23.
Al-Dhaheri M, Wu J, Skliris GP, Li J, Higashimato K, Wang Y, et al. CARM1 is an important determinant of ERalpha-dependent breast cancer cell differentiation and proliferation in breast cancer cells. Cancer Res. 2011;71:2118–28.
Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–6.
Asakawa H, Hiraoka Y, Haraguchi T. Estimation of GFP-Nucleoporin amount based on fluorescence microscopy. Methods Mol Biol. 2018;1721:105–15.
Rodriguez-Bravo V, Pippa R, Song WM, Carceles-Cordon M, Dominguez-Andres A, Fujiwara N, et al. Nuclear Pores Promote Lethal Prostate Cancer by Increasing POM121-Driven E2F1, MYC, and AR Nuclear Import. Cell. 2018;174:1200–15 e1220.
Zhou MH, Yang QM. NUP214 fusion genes in acute leukemia (Review). Oncol Lett. 2014;8:959–62.
Wang XM, Yao M, Liu SX, Hao J, Liu QJ, Gao F. Interplay between the Notch and PI3K/Akt pathways in high glucose-induced podocyte apoptosis. Am J Physiol Ren Physiol. 2014;306:F205–213.
Demitrack ES, Samuelson LC. Notch as a driver of gastric epithelial cell proliferation. Cell Mol Gastroenterol Hepatol. 2017;3:323–30.
Penton AL, Leonard LD, Spinner NB. Notch signaling in human development and disease. Semin Cell Developmental Biol. 2012;23:450–7.
Xiu MX, Liu YM. The role of oncogenic Notch2 signaling in cancer: A novel therapeutic target. Am J Cancer Res. 2019;9:837–54.
Kunnimalaiyaan S, Gamblin TC, Kunnimalaiyaan M. Glycogen synthase kinase-3 inhibitor AR-A014418 suppresses pancreatic cancer cell growth via inhibition of GSK-3-mediated Notch1 expression. HPB: Off J Int Hepato Pancreato Biliary Assoc. 2015;17:770–6.
Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol cell. 2004;16:509–20.
Takeuchi H, Haltiwanger RS. Role of glycosylation of Notch in development. Semin Cell Developmental Biol. 2010;21:638–45.
Takeuchi H, Haltiwanger RS. Significance of glycosylation in Notch signaling. Biochemical Biophysical Res Commun. 2014;453:235–42.
Deng Q, Hou J, Feng L, Lv A, Ke X, Liang H, et al. PHF19 promotes the proliferation, migration, and chemosensitivity of glioblastoma to doxorubicin through modulation of the SIAH1/beta-catenin axis. Cell Death Dis. 2018;9:1049.
Zhang D, Wang F, Pang Y, Ke XX, Zhu S, Zhao E, et al. Down-regulation of CHERP inhibits neuroblastoma cell proliferation and induces apoptosis through ER stress induction. Oncotarget. 2017;8:80956–70.
Wang F, Zhang D, Mao J, Ke XX, Zhang R, Yin C, et al. Morusin inhibits cell proliferation and tumor growth by down-regulating c-Myc in human gastric cancer. Oncotarget. 2017;8:57187–200.
Acknowledgements
This study was supported by the fellowship of China Postdoctoral Science Foundation (2021M692679), the special support from Chongqing Postdoctoral Research Institute (7820100607), the National Key Research, and Development Program of China (No.2016YFC1302204 and 2017YFC308600). We thank GEPIA 2 database (http://gepia2.cancer-pku.cn/#survival), TIMER 2.0 database (http://timer.cistrome.org/), Oncomine database (https://www.oncomine.org/resource/login.html) for initial data analysis. We are grateful to PMeS (http://bioinfo.ncu.edu.cn/inquiries_PMeS.aspx) for the prediction of protein methylation sites. Technical support of confocal microscopy from Lei Zhang is greatly appreciated.
Author information
Authors and Affiliations
Contributions
Feng Wang conceived the study, designed and performed the experiments, analyzed the data, and wrote the paper. HC provided suggestions for experiments, reviewed and edited the manuscript. ZC provided suggestions for experiments and assisted with vector construction. MNA and HT assisted with the western blot and ChIP experiments. ZD and SC performed the MS experiment and analyzed the results. JZ and YP performed mice injections. XK and WP performed cell culture, transfection, and infection.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Note: The authors declare that the content of the manuscript is original and that it has not been published or accepted for publication, either in whole or in part, in any form.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, F., Zhang, J., Tang, H. et al. Nup54-induced CARM1 nuclear importation promotes gastric cancer cell proliferation and tumorigenesis through transcriptional activation and methylation of Notch2. Oncogene 41, 246–259 (2022). https://doi.org/10.1038/s41388-021-02078-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02078-9
This article is cited by
-
The emerging role of CARM1 in cancer
Cellular Oncology (2024)
-
NCX1 coupled with TRPC1 to promote gastric cancer via Ca2+/AKT/β-catenin pathway
Oncogene (2022)