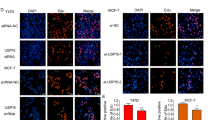
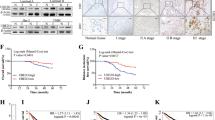
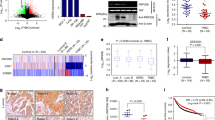
Abstract
ERα signaling drives proliferation, survival and cancer initiation in the mammary gland. Therefore, it is critical to elucidate mechanisms by which ERα expression is regulated. We show that the tumor suppressor E3 ligase COP1 promotes the degradative polyubiquitination of the microtubule-associated protein HPIP. As such, COP1 negatively regulates estrogen-dependent AKT activation in breast cancer cells. However, COP1 also induces ERα expression and ERα-dependent gene transcription, at least through c-Jun degradation. COP1 and ERα levels are positively correlated in clinical cases of breast cancer. COP1 also supports the metabolic reprogramming by estrogens, including glycolysis. On the other hand, COP1 suppresses EMT in breast cancer cells. COP1 deficiency also contributes to Tamoxifen resistance, at least through protective autophagy. Therefore, COP1 acts as an oncogenic E3 ligase by promoting ERα signaling but also acts as a tumor suppressor candidate by preventing EMT, which reflects a dual role of COP1 in breast cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
References
Craig Allred D, Brown P, Medina D. The origins of estrogen receptor alpha-positive and estrogen receptor alpha-negative human breast cancer. Breast Cancer Res. 2004;6:240–5.
Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME. GREB1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92:141–9.
Ikeda K, Horie-Inoue K, Inoue S. Identification of estrogen-responsive genes based on the DNA binding properties of estrogen receptors using high-throughput sequencing technology. Acta Pharmacol Sin. 2015;36:24–31.
Welboren WJ, Van Driel MA, Janssen-Megens EM, Van Heeringen SJ, Sweep FC, Span PN, et al. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–28.
Welboren WJ, Sweep FCGJ, Span PN, Stunnenberg HG. Genomic actions of estrogen receptor α: What are the targets and how are they regulated? Endocr-Relat Cancer. 2009;16:1073–89.
Folkerd EJ, Dowsett M. Influence of sex hormones on cancer progression. J Clin Oncol. 2010;28:4038–44.
Koš M, Reid G, Denger S, Gannon F. Minireview: genomic organization of the human ERα gene promoter region. Mol Endocrinol. 2001;15:2057–63.
Yoshida T, Eguchi H, Nakachi K, Tanimoto K, Higashi Y, Suemasu K, et al. Distinct mechanisms of loss of estrogen receptor α gene expression in human breast cancer: Methylation of the gene and alteration of trans-acting factors. Carcinogenesis. 2000;21:2193–201.
Turner BC, Zhang J, Gumbs AA, Maher MG, Kaplan L, Carter D, et al. Expression of AP-2 transcription factors in human breast cancer correlates with the regulation of multiple growth factor signalling pathways. Cancer Res. 1998;58:5466–72.
Grabinski N, Möllmann K, Milde-Langosch K, Müller V, Schumacher U, Brandt B, et al. AKT3 regulates ErbB2, ErbB3 and estrogen receptor α expression and contributes to endocrine therapy resistance of ErbB2+ breast tumor cells from Balb-neuT mice. Cell Signal. 2014;26:1021–9.
Madureira PA, Varshochi R, Constantinidou D, Francis RE, Coombes RC, Yao KM, et al. The forkhead box M1 protein regulates the transcription of the estrogen receptor α in breast cancer cells. J Biol Chem. 2006;281:25167–76.
Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor α expression in breast cancer. Cancer Res. 2007;67:6477–83.
Doucas V, Spyrou G, Yaniv M. Unregulated expression of c-Jun or c-Fos proteins but not Jun D inhibits oestrogen receptor activity in human breast cancer derived cells. EMBO J. 1991;10:2237–45.
Smith LM, Wise SC, Hendricks DT, Sabichi AL, Bos T, Reddy P, et al. cJun overexpression in MCF-7 breast cancer cells produces a tumorigenic, invasive and hormone resistant phenotype. Oncogene. 1999;18:6063–70.
Tecalco-Cruz AC, Ramírez-Jarquín JO. Mechanisms that increase stability of estrogen receptor alpha in breast cancer. Clin Breast Cancer. 2017;17:1–10.
Manavathi B, Acconcia F, Rayala SK, Kumar R. An inherent role of microtubule network in the action of nuclear receptor. Proc Natl Acad Sci USA. 2006;103:15981–6.
Shostak K, Patrascu F, Göktuna SI, Close P, Borgs L, Nguyen L, et al. MDM2 restrains estrogen-mediated AKT activation by promoting TBK1-dependent HPIP degradation. Cell Death Differ. 2014;21:811–24.
Marine JC. Spotlight on the role of COP1 in tumorigenesis. Nat Rev Cancer. 2012;12:455–64.
Migliorini D, Bogaerts S, Defever D, Vyas R, Denecker G, Radaelli E, et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J Clin Investig. 2011;121:1329–43.
Vitari AC, Leong KG, Newton K, Yee C, Oĝrourke K, Liu J, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474:403–8.
Baert JL, Monte D, Verreman K, Degerny C, Coutte L, De Launoit Y. The E3 ubiquitin ligase complex component COP1 regulates PEA3 group member stability and transcriptional activity. Oncogene. 2010;29:1810–20.
Wertz IE, O’Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, et al. Human De-etiolated-regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science. 2004;303:1371–4.
Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92.
Lorent J, Kusnadi EP, Hoef V, Rebello RJ, Leibovitch M, Ristau J, et al. Translational offsetting as a mode of estrogen receptor α‐dependent regulation of gene expression. EMBO J. 2019;38. https://doi.org/10.15252/embj.2018101323.
Neeman M, Degani H. Metabolic studies of estrogen-and tamoxifen-treated human breast cancer cells by nuclear magnetic resonance spectroscopy. Cancer Res. 1989;49:589–94.
Neeman M, Degani H. Early estrogen-induced metabolic changes and their inhibition by actinomycin D and cycloheximide in human breast cancer cells: 31P and 13C NMR studies. Proc Natl Acad Sci USA. 1989;86:5585–9.
O’Mahony F, Razandi M, Pedram A, Harvey BJ, Levin ER. Estrogen modulates metabolic pathway adaptation to available glucose in breast cancer cells. Mol Endocrinol. 2012;26:2058–70.
Jia M, Andreassen T, Jensen L, Bathen TF, Sinha I, Gao H, et al. Estrogen receptor a promotes breast cancer by reprogramming choline metabolism. Cancer Res. 2016;76:5634–46.
van Gastel N, Stegen S, Eelen G, Schoors S, Carlier A, Daniëls VW, et al. Lipid availability determines fate of skeletal progenitor cells via SOX9. Nature. 2020;579:111–7.
Shostak K, Zhang X, Hubert P, Göktuna SI, Jiang Z, Klevernic I, et al. NF-κB-induced KIAA1199 promotes survival through EGFR signalling. Nat Commun. 2014;5. https://doi.org/10.1038/ncomms6232.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates. Cells Prop Stem Cells Cell. 2008;133:704–15.
Luque-Bolivar A, Pérez-Mora E, Eugenia Villegas V, Rondon-Lagos M. Resistance and overcoming resistance in breast cancer. Breast Cancer. 2020;12:211–29.
Cook KL, Shajahan AN, Wärri A, Jin L, Hilakivi-Clarke LA, Clarke R. Glucose-regulated protein 78 controls cross-talk between apoptosis and autophagy to determine antiestrogen responsiveness. Cancer Res. 2012;72:3337–49.
Samaddar JS, Gaddy VT, Duplantier J, Thandavan SP, Shah M, Smith MJ, et al. A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol Cancer Ther. 2008;7:2977–87.
Meley D, Bauvy C, Houben-Weerts JHPM, Dubbelhuis PF, Helmond MTJ, Codogno P, et al. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–9.
Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41.
Chan EYW, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–74.
Zou S, Zhu Y, Wang B, Qian F, Zhang X, Wang L, et al. The ubiquitin ligase COP1 promotes glioma cell proliferation by preferentially downregulating tumor suppressor p53. Mol Neurobiol. 2017;54:5008–16.
Wang SC, Chai DSen, Chen CB, Wang ZY, Wang L. HPIP promotes thyroid cancer cell growth, migration and EMT through activating PI3K/AKT signaling pathway. Biomed Pharmacother. 2015;75:33–39.
Ouyang M, Wang H, Ma J, Lü W, Li J, Yao C, et al. COP1, the negative regulator of ETV1, influences prognosis in triple-negative breast cancer. BMC Cancer. 2015;15. https://doi.org/10.1186/s12885-015-1151-y.
Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–61.
Vega S, Morales AV, Ocaña OH, Valdés F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–43.
Hawley SA, Ross FA, Chevtzoff C, Green KA, Evans A, Fogarty S, et al. Use of cells expressing γ subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab. 2010;11:554–65.
Shostak K, Jiang Z, Charloteaux B, Mayer A, Habraken Y, Tharun L, et al. The X-linked trichothiodystrophy-causing gene RNF113A links the spliceosome to cell survival upon DNA damage. Nat Commun. 2020;11. https://doi.org/10.1038/s41467-020-15003-7.
Gatot JS, Gioia R, Chau TL, Patrascu F, Warnier M, Close P, et al. Lipopolysaccharide-mediated interferon regulatory factor activation involves TBK1-IKKε-dependent lys63-linked polyubiquitination and phosphorylation of TANK/I-TRAF. J Biol Chem. 2007;282:31131–46.
Hendriks MMWB, Smit S, Akkermans WLMW, Reijmers TH, Eilers PHC, Hoefsloot HCJ, et al. How to distinguish healthy from diseased? Classification strategy for mass spectrometry-based clinical proteomics. Proteomics. 2007;7:3672–80.
Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257.
Rademaker G, Hennequière V, Brohée L, Nokin MJ, Lovinfosse P, Durieux F, et al. Myoferlin controls mitochondrial structure and activity in pancreatic ductal adenocarcinoma, and affects tumor aggressiveness. Oncogene. 2018;37:4398–412.
Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38:276–8.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15. https://doi.org/10.1186/s13059-014-0550-8.
Acknowledgements
The authors thank Arnaud Blomme (Laboratory of Cancer Signaling, University of Liege, Liege, Belgium) for the gift of PyMT cells. The authors also thank the GIGA Imaging and Flow Cytometry Facility as well as the GIGA Genomics Platform for RNA-Sequencing analyses. This study was supported by Grants from the Belgian National Funds for Scientific Research (FNRS) and from Special Research Funds (FSR) at the University of Liege, the Belgian foundation against Cancer (FAF-F/2016/794), as well as from the Walloon Excellence in Life Sciences and Biotechnology (WELBIO-CR-2015A-02). We are also grateful to the “Fondation Leon Fredericq” of the CHU Liege for its financial support. A. Chariot and P. Close are Research Director and Senior Research Associate at the FNRS, respectively. The authors declare no conflict of interest.
Author information
Authors and Affiliations
Contributions
Conceptualization, AC and KS; Methodology, SCT, QL, OP, PCh, AL, A M, PAF, PCl, SK, AF and IN; Investigation, SCT, QL, PCh, AL, AM, PAF, PCl, SK, AF and IN; Formal Analysis: SCT, OP, KS and AC; Writing—Original Draft, AC; Funding Acquisition, AC; Resources, RB; Supervision, AC.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Tang, S.C., Lion, Q., Peulen, O. et al. The E3 ligase COP1 promotes ERα signaling and suppresses EMT in breast cancer. Oncogene 41, 173–190 (2022). https://doi.org/10.1038/s41388-021-02038-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02038-3
This article is cited by
-
Deubiquitinase YOD1 suppresses tumor progression by stabilizing E3 ligase TRIM33 in head and neck squamous cell carcinoma
Cell Death & Disease (2023)
-
The crosstalk between ubiquitination and endocrine therapy
Journal of Molecular Medicine (2023)