Abstract
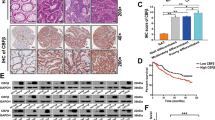
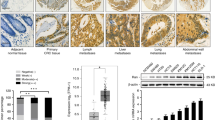
Colorectal cancer (CRC) is among the top five most common malignant tumors worldwide and has a high mortality rate. Identification of the mechanism of CRC and potential therapeutic targets is critical for improving survival. In the present study, we observed high expression of RAN binding protein 1 (RANBP1) in CRC tissues. Upregulated RANBP1 expression was strongly associated with TNM stages and was an independent risk factor for poor prognosis. In vitro and in vivo functional experiments demonstrated that RANBP1 promoted the proliferation and invasion of CRC cells and inhibited the apoptosis of CRC cells. Low RANBP1 expression reduced the expression levels of hsa-miR-18a, hsa-miR-183, and hsa-miR-106 microRNAs (miRNAs) by inhibiting the nucleoplasmic transport of precursor miRNAs (pre-miRNAs), thereby promoting the accumulation of the latter in the nucleus and reducing the expression of mature miRNAs. Further experiments and bioinformatic analyses demonstrated that RANBP1 promoted the expression of YAP by regulating miRNAs and the Hippo pathway. We also found that YAP acted as a transcriptional cofactor to activate RANBP1 transcription in combination with TEAD4 transcription factor. Thus, RANBP1 further promoted the progression of CRC by forming a positive feedback loop with YAP. Our results revealed the biological role and mechanism of RANBP1 in CRC for the first time, suggesting that RANBP1 can be used as a diagnostic molecule and a potential therapeutic target in CRC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
04 January 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41388-021-02152-2
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries (vol 68, pg 394, 2018). Ca-Cancer J Clin. 2020;70:313–313.
Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer commun. 2019;39:22.
Di Fiore B, Ciciarello M, Mangiacasale R, Palena A, Tassin AM, Cundari E, et al. Mammalian RanBP1 regulates centrosome cohesion during mitosis. J cell Sci. 2003;116:3399–411.
Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J cell Sci. 2008;121:1577–86.
Kehlenbach RH, Dickmanns A, Kehlenbach A, Guan TL, Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol. 1999;145:645–57.
Oh D, Yu CH, Needleman DJ. Spatial organization of the Ran pathway by microtubules in mitosis. Proc Natl Acad Sci USA. 2016;113:8729–34.
Zhang QM, Huang SS, Luo HR, Zhao X, Wu G, Wu DL. Eight-plex iTRAQ labeling and quantitative proteomic analysis for human bladder cancer. Am J cancer Res. 2017;7:935–45.
Rensen WM, Roscioli E, Tedeschi A, Mangiacasale R, Ciciarello M, Di Gioia SA, et al. RanBP1 downregulation sensitizes cancer cells to taxol in a caspase-3-dependent manner. Oncogene. 2009;28:1748–58.
Dattilo V, D’Antona L, Talarico C, Capula M, Catalogna G, Iuliano R, et al. SGK1 affects RAN/RANBP1/RANGAP1 via SP1 to play a critical role in pre-miRNA nuclear export: a new route of epigenomic regulation. Sci Rep. 2017;7:45361.
Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends cell Biol. 2004;14:156–9.
Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–57.
Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79.
Zhang L, Tang F, Terracciano L, Hynx D, Kohler R, Bichet S, et al. NDR functions as a physiological YAP1 kinase in the intestinal epithelium. Curr Biol: CB. 2015;25:296–305.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol-Mech. 2014;9:287–314.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66.
Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang QQ, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–U143.
Lu YY, Zhao XD, Liu Q, Li CX, Graves-Deal R, Cao Z, et al. IncRNA MIR100HG-derived miR-100 and miR-125b m ediate cetuximab resistance via Wnt/beta-catenin signaling. Nat Med. 2017;23:1331–+.
Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis (vol 464, pg 1196, 2010). Nature. 2010;467:356–356.
Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Bio. 2005;6:376–85.
Lin SB, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–33.
Lee Y, Kim M, Han JJ, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–60.
Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40.
Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5.
Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6.
Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98.
Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6.
Muralidhar B, Winder D, Murray M, Palmer R, Barbosa-Morais N, Saini H, et al. Functional evidence that Drosha overexpression in cervical squamous cell carcinoma affects cell phenotype and microRNA profiles. J Pathol. 2011;224:496–507.
Shu GS, Yang ZL, Liu DC. Immunohistochemical study of Dicer and Drosha expression in the benign and malignant lesions of gallbladder and their clinicopathological significances. Pathol Res Pr. 2012;208:392–7.
Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–4.
Wagh PK, Gardner MA, Ma XL, Callahan M, Shannon JM, Wert SE, et al. Cell-and developmental stage-specific Dicer1 ablation in the lung epithelium models cystic pleuropulmonary blastoma. J Pathol. 2015;236:41–52.
Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A genetic defect in exportin-5 traps precursor micrornas in the nucleus of cancer cells. Cancer cell. 2010;18:303–15.
Shigeyasu K, Okugawa Y, Toden S, Boland CR, Goel A. Exportin-5 functions as an oncogene and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23:1312–22.
Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15:642–56.
Zhou QW, Hou ZB, Zuo SY, Zhou X, Feng YD, Sun Y, et al. LUCAT1 promotes colorectal cancer tumorigenesis by targeting the ribosomal protein L40-MDM2-p53 pathway through binding with UBA52. Cancer Sci. 2019;110:1194–207.
Detre S, Saclani Jotti G, Dowsett M. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–8.
Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PloS one. 2012;7:e51862.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 81972288) and the General Project of the Natural Science Foundation of Jiangsu Province (no. BK20191353).
Author information
Authors and Affiliations
Contributions
XY designed the study and wrote the manuscript. DZ and SZ developed the methodology and performed the analyses. MC and CZ collected and analyzed the clinical data. XX performed the statistical analysis. YL and SD performed the bioinformatics analysis. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zheng, D., Cao, M., Zuo, S. et al. RANBP1 promotes colorectal cancer progression by regulating pre-miRNA nuclear export via a positive feedback loop with YAP. Oncogene 41, 930–942 (2022). https://doi.org/10.1038/s41388-021-02036-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-02036-5
This article is cited by
-
The interplay between noncoding RNA and YAP/TAZ signaling in cancers: molecular functions and mechanisms
Journal of Experimental & Clinical Cancer Research (2022)