Abstract
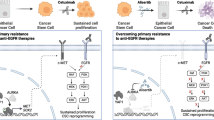
The emergence of RAS/RAF mutant clone is the main feature of EGFR inhibitor resistance in KRAS wild-type colon cancer. However, its molecular mechanism is thought to be multifactorial, mainly due to cellular heterogeneity. In order to better understand the resistance mechanism in a single clone level, we successfully isolated nine cells with cetuximab-resistant (CR) clonality from in vitro system. All CR cells harbored either KRAS or BRAF mutations. Characteristically, these cells showed a higher EMT (Epithelial to mesenchymal transition) signature, showing increased EMT markers such as SNAI2. Moreover, the expression level of CXCL1/5, a secreted protein, was significantly higher in CR cells compared to the parental cells. In these CR cells, CXCL1/5 expression was coordinately regulated by SNAI2/NFKB and transactivated EGFR through CXCR/MMPI/EGF axis via autocrine singling. We also observed that combined cetuximab/MEK inhibitor not only showed growth inhibition but also reduced the secreted amounts of CXCL1/5. We further found that serum CXCL1/5 level was positively correlated with the presence of RAS/RAF mutation in colon cancer patients during cetuximab therapy, suggesting its role as a biomarker. These data indicated that the application of serum CXCL1/5 could be a potential serologic biomarker for predicting resistance to EGFR therapy in colorectal cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–93.
Mishra J, Drummond J, Quazi SH, Karanki SS, Shaw JJ, Chen B, et al. Prospective of colon cancer treatments and scope for combinatorial approach to enhanced cancer cell apoptosis. Crit Rev Oncol Hematol. 2013;86:232–50.
Cremolini C, Schirripa M, Antoniotti C, Moretto R, Salvatore L, Masi G, et al. First-line chemotherapy for mCRC-a review and evidence-based algorithm. Nat Rev Clin Oncol. 2015;12:607–19.
Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–74.
Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81–94.
Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014;4:1269–80.
Hsu HC, Thiam TK, Lu YJ, Yeh CY, Tsai WS, You JF, et al. Mutations of KRAS/NRAS/BRAF predict cetuximab resistance in metastatic colorectal cancer patients. Oncotarget. 2016;7:22257–70.
Misale S, Arena S, Lamba S, Siravegna G, Lallo A, Hobor S, et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci Transl Med. 2014;6:224ra226.
Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6.
van Brummelen EMJ, de Boer A, Beijnen JH, Schellens JHM. BRAF mutations as predictive biomarker for response to anti-EGFR monoclonal antibodies. Oncologist. 2017;22:864–72.
Zhao B, Wang L, Qiu H, Zhang M, Sun L, Peng P, et al. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget. 2017;8:3980–4000.
Arena S, Bellosillo B, Siravegna G, Martinez A, Canadas I, Lazzari L, et al. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin Cancer Res. 2015;21:2157–66.
Xu JM, Wang Y, Wang YL, Wang Y, Liu T, Ni M, et al. PIK3CA mutations contribute to acquired cetuximab resistance in patients with metastatic colorectal cancer. Clin Cancer Res. 2017;23:4602–16.
Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat Rev Cancer. 2004;4:11–22.
Ramesh G, MacLean AG, Philipp MT. Cytokines and chemokines at the crossroads of neuroinflammation, neurodegeneration, and neuropathic pain. Mediators Inflamm. 2013;2013:480739.
Turner MD, Nedjai B, Hurst T, Pennington DJ. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta. 2014;1843:2563–82.
Bolitho C, Hahn MA, Baxter RC, Marsh DJ. The chemokine CXCL1 induces proliferation in epithelial ovarian cancer cells by transactivation of the epidermal growth factor receptor. Endocr Relat Cancer. 2010;17:929–40.
Grepin R, Guyot M, Giuliano S, Boncompagni M, Ambrosetti D, Chamorey E, et al. The CXCL7/CXCR1/2 axis is a key driver in the growth of clear cell renal cell carcinoma. Cancer Res. 2014;74:873–83.
Kuo PL, Shen KH, Hung SH, Hsu YL. CXCL1/GROalpha increases cell migration and invasion of prostate cancer by decreasing fibulin-1 expression through NF-kappaB/HDAC1 epigenetic regulation. Carcinogenesis. 2012;33:2477–87.
Spaks A. Role of CXC group chemokines in lung cancer development and progression. J Thorac Dis. 2017;9:S164–S171.
Sparmann A, Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell. 2004;6:447–58.
Yuan M, Zhu H, Xu J, Zheng Y, Cao X, Liu Q. Tumor-derived CXCL1 promotes lung cancer growth via recruitment of tumor-associated neutrophils. J Immunol Res. 2016;2016:6530410.
Kogan-Sakin I, Cohen M, Paland N, Madar S, Solomon H, Molchadsky A, et al. Prostate stromal cells produce CXCL-1, CXCL-2, CXCL-3 and IL-8 in response to epithelia-secreted IL-1. Carcinogenesis. 2009;30:698–705.
Mangahas CR, dela Cruz GV, Friedman-Jimenez G, Jamal S. Endothelin-1 induces CXCL1 and CXCL8 secretion in human melanoma cells. J Investig Dermatol. 2005;125:307–11.
Jones VS, Huang RY, Chen LP, Chen ZS, Fu L, Huang RP. Cytokines in cancer drug resistance: Cues to new therapeutic strategies. Biochim Biophys Acta. 2016;1865:255–65.
Nolen BM, Marks JR, Ta’san S, Rand A, Luong TM, Wang Y, et al. Serum biomarker profiles and response to neoadjuvant chemotherapy for locally advanced breast cancer. Breast Cancer Res. 2008;10:R45.
Pommier AJ, Shaw R, Spencer SK, Morgan SR, Hoff PM, Robertson JD, et al. Serum protein profiling reveals baseline and pharmacodynamic biomarker signatures associated with clinical outcome in mCRC patients treated with chemotherapy +/− cediranib. Br J Cancer. 2014;111:1590–604.
Shaw VE, Lane B, Jenkinson C, Cox T, Greenhalf W, Halloran CM, et al. Serum cytokine biomarker panels for discriminating pancreatic cancer from benign pancreatic disease. Mol Cancer. 2014;13:114.
Yako YY, Kruger D, Smith M, Brand M. Cytokines as biomarkers of pancreatic ductal adenocarcinoma: a systematic review. PLoS ONE. 2016;11:e0154016.
Yamazaki N, Kiyohara Y, Uhara H, Iizuka H, Uehara J, Otsuka F, et al. Cytokine biomarkers to predict antitumor responses to nivolumab suggested in a phase 2 study for advanced melanoma. Cancer Sci. 2017;108:1022–31.
De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62.
Chen C, Xu ZQ, Zong YP, Ou BC, Shen XH, Feng H, et al. CXCL5 induces tumor angiogenesis via enhancing the expression of FOXD1 mediated by the AKT/NF-kappaB pathway in colorectal cancer. Cell Death Dis. 2019;10:178.
Hu B, Fan H, Lv X, Chen S, Shao Z. Prognostic significance of CXCL5 expression in cancer patients: a meta-analysis. Cancer Cell Int. 2018;18:68.
Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Mol Cancer. 2013;12:86.
Park MH, Hong JT. Roles of NF-kappaB in cancer and inflammatory diseases and their therapeutic approaches. Cells. 2016;5:15.
Xia Y, Shen S, Verma IM. NF-kappaB, an active player in human cancers. Cancer Immunol Res. 2014;2:823–30.
Bommarito A, Richiusa P, Carissimi E, Pizzolanti G, Rodolico V, Zito G, et al. BRAFV600E mutation, TIMP-1 upregulation, and NF-kappaB activation: closing the loop on the papillary thyroid cancer trilogy. Endocr Relat Cancer. 2011;18:669–85.
Burke SJ, Lu D, Sparer TE, Masi T, Goff MR, Karlstad MD, et al. NF-kappaB and STAT1 control CXCL1 and CXCL2 gene transcription. Am J Physiol Endocrinol Metab. 2014;306:E131–149.
Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, et al. KrasG12D-induced IKK2/beta/NF-kappaB activation by IL-1alpha and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:105–20.
Mizumoto Y, Kyo S, Kiyono T, Takakura M, Nakamura M, Maida Y, et al. Activation of NF-kappaB is a novel target of KRAS-induced endometrial carcinogenesis. Clin Cancer Res. 2011;17:1341–50.
Grisanti LA, Guo S, Tilley DG. Cardiac GPCR-Mediated EGFR Transactivation: Impact and Therapeutic Implications. J Cardiovasc Pharmacol. 2017;70:3–9.
Wang Z. Transactivation of epidermal growth factor receptor by G protein-coupled receptors: recent progress, challenges and future research. Int J Mol Sci. 2016;17:95.
Milne GW, Nicklaus MC, Driscoll JS, Wang S, Zaharevitz D. National cancer institute drug information system 3D database. J Chem Inf Comput Sci. 1994;34:1219–24.
Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, et al. Metabolic heterogeneity in human lung tumors. Cell. 2016;164:681–94.
Saunders NA, Simpson F, Thompson EW, Hill MM, Endo-Munoz L, Leggatt G, et al. Role of intratumoural heterogeneity in cancer drug resistance: molecular and clinical perspectives. EMBO Mol Med. 2012;4:675–84.
Choi YH, Kim JK. Dissecting cellular heterogeneity using single-cell RNA sequencing. Mol Cells. 2019;42:189–99.
Goldman SL, MacKay M, Afshinnekoo E, Melnick AM, Wu S, Mason CE. The impact of heterogeneity on single-cell sequencing. Front Genet. 2019;10:8.
Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N, Werb Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat Cell Biol. 2018;20:1349–60.
Girbl T, Lenn T, Perez L, Rolas L, Barkaway A, Thiriot A, et al. Distinct compartmentalization of the chemokines CXCL1 and CXCL2 and the atypical receptor ACKR1 determine discrete stages of neutrophil diapedesis. Immunity. 2018;49:1062–76.e1066.
Sokol CL, Luster AD. The chemokine system in innate immunity. Cold Spring Harb Perspect Biol. 2015;7:a016303.
Susek KH, Karvouni M, Alici E, Lundqvist A. The role of CXC chemokine receptors 1-4 on immune cells in the tumor microenvironment. Front Immunol. 2018;9:2159.
Park YL, Kim HP, Cho YW, Min DW, Cheon SK, Lim YJ, et al. Activation of WNT/beta-catenin signaling results in resistance to a dual PI3K/mTOR inhibitor in colorectal cancer cells harboring PIK3CA mutations. Int J Cancer. 2019;144:389–401.
Yun J, Song SH, Kang JY, Park J, Kim HP, Han SW, et al. Reduced cohesin destabilizes high-level gene amplification by disrupting pre-replication complex bindings in human cancers with chromosomal instability. Nucleic Acids Res. 2016;44:558–72.
Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007;28:810–22.
Taylor SC, Laperriere G, Germain H. Droplet digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci Rep. 2017;7:2409.
Acknowledgements
We thank H. Nikki March, Ph.D., from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript. The authors are grateful to all members of our group for their helpful advice. This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare (HI14C1277, HI13C2096), by the Ministry of Science, ICT & Future Planning (2016M3A9B6026918, 2018R1C1B6005264), and by the National Research Foundation of Korea (2017M3C9A5029978), and by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science & ICT (NRF-2017M3A9A7050610).
Author information
Authors and Affiliations
Contributions
Y-LP, H-PK, and T-YK conceived the project; Y-LP, D-WM established xenograft models Y-LP performed single-cell experiments; C-YO and JKK performed computational analysis; Y-LP designed and performed in vitro and in vivo experiments; Y-LP, H-PK, YJL, S-HS, S-WH, and T-YK interpreted data; Y-LP wrote the manuscript. All authors contributed edits and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Park, YL., Kim, HP., Ock, CY. et al. EMT-mediated regulation of CXCL1/5 for resistance to anti-EGFR therapy in colorectal cancer. Oncogene 41, 2026–2038 (2022). https://doi.org/10.1038/s41388-021-01920-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01920-4