Abstract
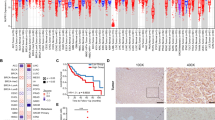
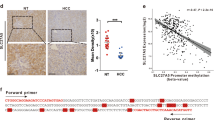
SWItch/Sucrose Non-Fermentable (SWI/SNF) is a multiprotein complex essential for the regulation of eukaryotic gene expression. SWI/SNF complex genes are genetically altered in over 20% of human malignancies, but the aberrant regulation of the SWI/SNF subunit genes and subsequent dysfunction caused by abnormal expression of subunit gene in cancer, remain poorly understood. Among the SWI/SNF subunit genes, SMARCA4, SMARCC1, and SMARCA2 were identified to be overexpressed in human hepatocellular carcinoma (HCC). Modulation of SMARCA4, SMARCC1, and SMARCA2 inhibited in vitro tumorigenesis of HCC cells. However, SMARCA4-targeting elicited remarkable inhibition in an in vivo Ras-transgenic mouse HCC model (Ras-Tg), and high expression levels of SMARCA4 significantly associated with poor prognosis in HCC patients. Furthermore, most HCC patients (72–86%) showed SMARCA4 overexpression compared to healthy controls. To identify SMARCA4-specific active enhancers, mapping, and analysis of chromatin state in liver cancer cells were performed. Integrative analysis of SMARCA4-regulated genes and active chromatin enhancers suggested 37 genes that are strongly activated by SMARCA4 in HCC. Through chromatin immunoprecipitation-qPCR and luciferase assays, we demonstrated that SMARCA4 activates Interleukin-1 receptor-associated kinase 1 (IRAK1) expression through IRAK1 active enhancer in HCC. We then showed that transcriptional activation of IRAK1 induces oncoprotein Gankyrin and aldo–keto reductase family 1 member B10 (AKR1B10) in HCC. The regulatory mechanism of the SMARCA4-IRAK1-Gankyrin, AKR1B10 axis was further demonstrated in HCC cells and in vivo Ras-Tg mice. Our results suggest that aberrant overexpression of SMARCA4 causes SWI/SNF to promote IRAK1 enhancer to activate oncoprotein Gankyrin and AKR1B10, thereby contributing to hepatocarcinogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Michel BC, D’Avino AR, Cassel SH, Mashtalir N, McKenzie ZM, McBride MJ, et al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat Cell Biol. 2018;20:1410–20.
Mashtalir N, D’Avino AR, Michel BC, Luo J, Pan J, Otto JE. et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell. 2018;175:1272–88.e1220.
Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601.
Jubierre L, Soriano A, Planells-Ferrer L, Paris-Coderch L, Tenbaum SP, Romero OA, et al. BRG1/SMARCA4 is essential for neuroblastoma cell viability through modulation of cell death and survival pathways. Oncogene. 2016;35:5179–90.
Buscarlet M, Krasteva V, Ho L, Simon C, Hebert J, Wilhelm B, et al. Essential role of BRG, the ATPase subunit of BAF chromatin remodeling complexes, in leukemia maintenance. Blood. 2014;123:1720–8.
Sun X, Wang SC, Wei Y, Luo X, Jia Y, Li L, et al. Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell. 2017;32:574–89 e576.
Wang P, Song X, Cao D, Cui K, Wang J, Utpatel K, et al. Oncogene-dependent function of BRG1 in hepatocarcinogenesis. Cell Death Dis. 2020;11:91.
O’Neil NJ, Bailey ML, Hieter P. Synthetic lethality and cancer. Nat Rev Genet. 2017;18:613–23.
Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–86.
Alver BH, Kim KH, Lu P, Wang X, Manchester HE, Wang W, et al. The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun. 2017;8:14648.
Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49:296–302.
Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49.
Su B, Luo T, Zhu J, Fu J, Zhao X, Chen L, et al. Interleukin-1beta/Iinterleukin-1 receptor-associated kinase 1 inflammatory signaling contributes to persistent Gankyrin activation during hepatocarcinogenesis. Hepatology. 2015;61:585–97.
Cheng BY, Lau EY, Leung HW, Leung CO, Ho NP, Gurung S, et al. IRAK1 augments cancer stemness and drug resistance via the AP-1/AKR1B10 signaling cascade in hepatocellular carcinoma. Cancer Res. 2018;78:2332–42.
Saladi SV, Ross K, Karaayvaz M, Tata PR, Mou H, Rajagopal J, et al. ACTL6A is co-amplified with p63 in squamous cell carcinoma to drive YAP Activation, Regenerative Proliferation, and Poor Prognosis. Cancer Cell. 2017;31:35–49.
Hu B, Lin JZ, Yang XB, Sang XT. The roles of mutated SWI/SNF complexes in the initiation and development of hepatocellular carcinoma and its regulatory effect on the immune system: A review. Cell Prolif. 2020;53:e12791.
Cao D, Fan ST, Chung SS. Identification and characterization of a novel human aldose reductase-like gene. J Biol Chem. 1998;273:11429–35.
Wang AG, Moon HB, Lee MR, Hwang CY, Kwon KS, Yu SL, et al. Gender-dependent hepatic alterations in H-ras12V transgenic mice. J Hepatol. 2005;43:836–44.
Yang HD, Kim HS, Kim SY, Na MJ, Yang G, Eun JW, et al. HDAC6 suppresses Let-7i-5p to elicit TSP1/CD47-mediated anti-tumorigenesis and phagocytosis of hepatocellular carcinoma. Hepatology. 2019;70:1262–79.
Acknowledgements
We acknowledge support from the National Research Foundation (NRF) of Korea (2017R1A2B3002989 and 2021M3E5E7021893).
Author information
Authors and Affiliations
Contributions
Study conception and design: SYK, QS and SWN Acquisition of data: SYK, QS Analysis and interpretation of data: SYK, QS, HSK, HDY, MJN, ES, SY, KK, JSY, KY, SMJ, EKL, YMA, and WSP and Drafting of the paper: SWN. Critical revision of the paper: all authors. Technical and material support: SWN. Study supervision: SWN.
Corresponding author
Ethics declarations
Conflict of interest
SWN is the chief execute officer of NEORNAT, Inc. HSK and HDY are employee of NEORNAT, Inc. The other authors declare no conflict of interest relating to this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, S.Y., Shen, Q., Son, K. et al. SMARCA4 oncogenic potential via IRAK1 enhancer to activate Gankyrin and AKR1B10 in liver cancer. Oncogene 40, 4652–4662 (2021). https://doi.org/10.1038/s41388-021-01875-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01875-6