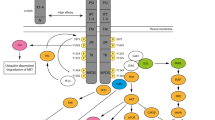
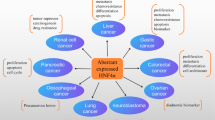
Abstract
This review provides a comprehensive landscape of HGF/c-MET (hepatocyte growth factor (HGF) /mesenchymal–epithelial transition factor (c-MET)) signaling pathway in cancers. First, we generalize the compelling influence of HGF/c-MET pathway on multiple cellular processes. Then, we present the genomic characterization of HGF/c-MET pathway in carcinogenesis. Furthermore, we extensively illustrate the malignant biological behaviors of HGF/c-MET pathway in cancers, in which hyperactive HGF/c-MET signaling is considered as a hallmark. In addition, we investigate the current clinical trials of HGF/c-MET-targeted therapy in cancers. We find that although HGF/c-MET-targeted therapy has led to breakthroughs in certain cancers, monotherapy of targeting HGF/c-MET has failed to demonstrate significant clinical efficacy in most cancers. With the advantage of the combinations of HGF/c-MET-targeted therapy, the exploration of more options of combinational targeted therapy in cancers may be the major challenge in the future.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Nakamura T, Nawa K, Ichihara A. Partial purification and characterization of hepatocyte growth factor from serum of hepatectomized rats. Biochem Biophys Res Commun. 1984;122:1450–9.
Thaler FJ, Michalopoulos GK. Hepatopoietin A: partial characterization and trypsin activation of a hepatocyte growth factor. Cancer Res. 1985;45:2545–9.
Zarnegar R, Michalopoulos G. Purification and biological characterization of human hepatopoietin A, a polypeptide growth factor for hepatocytes. Cancer Res. 1989;49:3314–20.
Nakamura T, Nishizawa T, Hagiya M, Seki T, Shimonishi M, Sugimura A, et al. Molecular cloning and expression of human hepatocyte growth factor. Nature. 1989;342:440–3.
Laguda B, Selden C, Jones M, Hodgson H, Spurr NK. Assignment of the hepatocyte growth factor (HGF) to chromosome 7q22-qter. Ann Hum Genet. 1991;55:213–6.
Stoker M, Gherardi E, Perryman M, Gray J. Scatter factor is a fibroblast-derived modulator of epithelial cell mobility. Nature. 1987;327:239–42.
Gherardi E, Sharpe M, Lane K, Sirulnik A, Stoker M. Hepatocyte growth factor/scatter factor (HGF/SF), the c-Met receptor and the behaviour of epithelial cells. Symp Soc Exp Biol. 1993;47:163–81.
Weidner KM, Behrens J, Vandekerckhove J, Birchmeier W. Scatter factor: molecular characteristics and effect on the invasiveness of epithelial cells. J Cell Biol. 1990;111:2097–108.
Gille J, Khalik M, König V, Kaufmann R. Hepatocyte growth factor/scatter factor (HGF/SF) induces vascular permeability factor (VPF/VEGF) expression by cultured keratinocytes. J Invest Dermatol. 1998;111:1160–5.
Higashio K, Shima N, Goto M, Itagaki Y, Nagao M, Yasuda H, et al. Identity of a tumor cytotoxic factor from human fibroblasts and hepatocyte growth factor. Biochem Biophys Res Commun. 1990;170:397–404.
Rubin JS, Chan AM, Bottaro DP, Burgess WH, Taylor WG, Cech AC, et al. A broad-spectrum human lung fibroblast-derived mitogen is a variant of hepatocyte growth factor. Proc Natl Acad Sci USA. 1991;88:415–9.
Schultz JM, Khan SN, Ahmed ZM, Riazuddin S, Waryah AM, Chhatre D, et al. Noncoding mutations of HGF are associated with nonsyndromic hearing loss, DFNB39. Am J Hum Genet. 2009;85:25–39.
Morell RJ, Olszewski R, Tona R, Leitess S, Wafa TT, Taukulis I, et al. Noncoding microdeletion in mouse Hgf disrupts neural crest migration into the stria vascularis, reduces the endocochlear potential and suggests the neuropathology for human nonsyndromic deafness DFNB39. J Neurosci. 2020;40:2976–92.
Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311:29–33.
Dean M, Park M, Le Beau MM, Robins TS, Diaz MO, Rowley JD, et al. The human met oncogene is related to the tyrosine kinase oncogenes. Nature. 1985;318:385–8.
Park M, Dean M, Kaul K, Braun MJ, Gonda MA, Vande, et al. Sequence of MET protooncogene cDNA has features characteristic of the tyrosine kinase family of growth-factor receptors. Proc Natl Acad Sci USA. 1987;84:6379–83.
Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, et al. Identification of the hepatocyte growth factor receptor as the c-Met proto-oncogene product. Science. 1991;251(Feb):802–4.
Jücker M, Günther A, Gradl G, Fonatsch C, Krueger G, Diehl V, et al. The Met/hepatocyte growth factor receptor (HGFR) gene is overexpressed in some cases of human leukemia and lymphoma. Leuk Res. 1994;18:7–16.
Mujtaba G, Schultz JM, Imtiaz A, Morell RJ, Friedman TB, Naz S. A mutation of MET, encoding hepatocyte growth factor receptor, is associated with human DFNB97 hearing loss. J Med Genet. 2015;52:548–52.
Gandino L, Longati P, Medico E, Prat M, Comoglio PM. Phosphorylation of ser 985 negatively regulates the hepatocyte growth factor receptor kinase. J Biol Chem. 1994;269:1815–20.
Kadoyama K, Funakoshi H, Ohya-Shimada W, Nakamura T, Matsumoto K, Matsuyama S, et al. Disease-dependent reciprocal phosphorylation of serine and tyrosine residues of c-Met/HGF receptor contributes disease retardation of a transgenic mouse model of ALS. Neurosci Res. 2009;65:194–200.
Nakayama M, Sakai K, Yamashita A, Nakamura T, Suzuki Y, Matsumoto K. Met/HGF receptor activation is regulated by juxtamembrane Ser985 phosphorylation in hepatocytes. Cytokine. 2013;62:446–52.
Peschard P, Fournier TM, Lamorte L, Naujokas MA, Band H, Langdon WY, et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol Cell. 2001;8:995–1004.
Rodrigues GA, Park M. Autophosphorylation modulates the kinase activity and oncogenic potential of the Met receptor tyrosine kinase. Oncogene. 1994;9:2019–27.
Suzuki M, Shiraha H, Fujikawa T, Takaoka N, Ueda N, Nakanishi Y, et al. Des-gamma-carboxy prothrombin is a potential autologous growth factor for hepatocellular carcinoma. J Biol Chem. 2005;280:6409–15.
Fan G, Zhang S, Gao Y, Greer PA, Tonks NK. HGF-independent regulation of MET and GAB1 by nonreceptor tyrosine kinase FER potentiates metastasis in ovarian cancer. Genes Dev. 2016;30:1542–57.
Corso S, Giordano S. Cell-autonomous and non-cell-autonomous mechanisms of HGF/MET-driven resistance to targeted therapies: from basic research to a clinical perspective. Cancer Discov. 2013;3:978–92.
Conrotto P, Corso S, Gamberini S, Comoglio PM, Giordano S. Interplay between scatter factor receptors and B plexins controls invasive growth. Oncogene. 2004;23:5131–7.
Chang L, Hu Y, Fu Y, Zhou T, You J, Du J, et al. Targeting slug-mediated non-canonical activation of c-Met to overcome chemo-resistance in metastatic ovarian cancer cells. Acta Pharm Sin B. 2019;9:484–95.
Wang H, Rao B, Lou J, Li J, Liu Z, Li A, et al. The function of the HGFc-Met axis in hepatocellular carcinoma. Front Cell Dev Biol. 2020;8:55.
Schmidt L, Duh FM, Chen F, Kishida T, Glenn G, Choyke P, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16:68–73.
Ma PC, Jagadeeswaran R, Jagadeesh S, Tretiakova MS, Nallasura V, Fox EA, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 2005;65:1479–88.
Ma PC, Kijima T, Maulik G, Fox EA, Sattler M, Griffin JD, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003;63:6272–81.
Krishnaswamy S, Kanteti R, Duke-Cohan JS, Loganathan S, Liu W, Ma PC, et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res. 2009;15:5714–23.
Lee JH, Han SU, Cho H, Jennings B, Gerrard B, Dean M, et al. A novel germ line juxtamembrane Met mutation in human gastric cancer. Oncogene. 2000;19:4947–53.
Ma PC, Tretiakova MS, MacKinnon AC, Ramnath N, Johnson C, Dietrich S, et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer. 2008;47:1025–37.
Puri N, Ahmed S, Janamanchi V, Tretiakova M, Zumba O, Krausz T, et al. Met is a potentially new therapeutic target for treatment of human melanoma. Clin Cancer Res. 2007;13:2246–53.
Jagadeeswaran R, Ma PC, Seiwert TY, Jagadeeswaran S, Zumba O, Nallasura V, et al. Functional analysis of c-Met/hepatocyte growth factor pathway in malignant pleural mesothelioma. Cancer Res. 2006;66:352–61.
Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66:283–9.
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50.
Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, Shivdasani P, et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J Clin Oncol. 2016;34:721–30.
Gow CH, Hsieh MS, Wu SG, Shih JY. A comprehensive analysis of clinical outcomes in lung cancer patients harboring a MET exon 14 skipping mutation compared to other driver mutations in an East Asian population. Lung Cancer. 2017;103:82–89.
Liu X, Jia Y, Stoopler MB, Shen Y, Cheng H, Chen J, et al. Next-generation sequencing of pulmonary sarcomatoid carcinoma reveals high frequency of actionable MET gene mutations. J Clin Oncol. 2016;34:794–802.
Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850–9.
Navis AC, van Lith SA, van Duijnhoven SM, de Pooter M, Yetkin-Arik B, Wesseling P, et al. Identification of a novel MET mutation in high-grade glioma resulting in an auto-active intracellular protein. Acta Neuropathol. 2015;130:131–44.
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8.
Asaoka Y, Tada M, Ikenoue T, Seto M, Imai M, Miyabayashi K, et al. Gastric cancer cell line Hs746T harbors a splice site mutation of c-Met causing juxtamembrane domain deletion. Biochem Biophys Res Commun. 2010;394:1042–6.
Umeki K, Shiota G, Kawasaki H. Clinical significance of c-Met oncogene alterations in human colorectal cancer. Oncology. 1999;56:314–21.
Tong CY, Hui AB, Yin XL, Pang JC, Zhu XL, Poon WS, et al. Detection of oncogene amplifications in medulloblastomas by comparative genomic hybridization and array-based comparative genomic hybridization. J Neurosurg. 2004;100:187–93.
Soman NR, Correa P, Ruiz BA, Wogan GN. The TPR-MET oncogenic rearrangement is present and expressed in human gastric carcinoma and precursor lesions. Proc Natl Acad Sci USA. 1991;88:4892–6.
Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846.
Bao ZS, Chen HM, Yang MY, Zhang CB, Yu K, Ye WL, et al. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res. 2014;24:1765–73.
International Cancer Genome Consortium PedBrain Tumor Project. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat Med. 2016;22:1314–20.
Kong LR, Mohamed Salleh NAB, Ong RW, Tan TZ, Syn NL, Goh RM, et al. A common MET polymorphism harnesses HER2 signaling to drive aggressive squamous cell carcinoma. Nat Commun. 2020;11:1556.
Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43.
Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19:495–509.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Xie Q, Bradley R, Kang L, Koeman J, Ascierto ML, Worschech A, et al. Hepatocyte growth factor (HGF) autocrine activation predicts sensitivity to MET inhibition in glioblastoma. Proc Natl Acad Sci USA. 2012;109:570–5.
Cheng F, Guo D. MET in glioma: signaling pathways and targeted therapies. J Exp Clin Cancer Res. 2019;38:270.
Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci USA. 2006;103:2316–21.
Pennacchietti S, Michieli P, Galluzzo M, Mazzone M, Giordano S, Comoglio PM. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 2003;3:347–61.
Daniel SK, Sullivan KM, Labadie KP, Pillarisetty VG. Hypoxia as a barrier to immunotherapy in pancreatic adenocarcinoma. Clin Transl Med. 2019;8:10.
Goyal L, Muzumdar MD, Zhu AX. Targeting the HGF/c-MET pathway in hepatocellular carcinoma. Clin Cancer Res. 2013;19:2310–8.
Tavian D, De Petro G, Benetti A, Portolani N, Giulini SM, Barlati S. u-PA and c-MET mRNA expression is co-ordinately enhanced while hepatocyte growth factor mRNA is down-regulated in human hepatocellular carcinoma. Int J Cancer. 2000;87:644–9.
Wu F, Wu L, Zheng S, Ding W, Teng L, Wang Z, et al. The clinical value of hepatocyte growth factor and its receptor-c-Met for liver cancer patients with hepatectomy. Dig Liver Dis. 2006;38:490–7.
Kondo S, Ojima H, Tsuda H, Hashimoto J, Morizane C, Ikeda M, et al. Clinical impact of c-Met expression and its gene amplification in hepatocellular carcinoma. Int J Clin Oncol. 2013;18:207–13.
Lee SJ, Lee J, Sohn I, Mao M, Kai W, Park CK, et al. A survey of c-MET expression and amplification in 287 patients with hepatocellular carcinoma. Anticancer Res. 2013;33:5179–86.
Harding JJ, Nandakumar S, Armenia J, Khalil DN, Albano M, Ly M, et al. Prospective genotyping of hepatocellular carcinoma: clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin Cancer Res. 2019;25:2116–26.
Nault JC, Martin Y, Caruso S, Hirsch TZ, Bayard Q, Calderaro J, et al. Clinical impact of genomic diversity from early to advanced hepatocellular carcinoma. Hepatology. 2020;71:164–82.
Davis IJ, McFadden AW, Zhang Y, Coxon A, Burgess TL, Wagner AJ, et al. Identification of the receptor tyrosine kinase c-Met and its ligand, hepatocyte growth factor, as therapeutic targets in clear cell sarcoma. Cancer Res. 2010;70:639–45.
Schöffski P, Wozniak A, Stacchiotti S, Rutkowski P, Blay JY, Lindner LH, et al. Activity and safety of crizotinib in patients with advanced clear-cell sarcoma with MET alterations: European Organization for Research and Treatment of Cancer phase II trial 90101 ‘CREATE’. Ann Oncol. 2017;28:3000–8.
Taulli R, Scuoppo C, Bersani F, Accornero P, Forni PE, Miretti S, et al. Validation of met as a therapeutic target in alveolar and embryonal rhabdomyosarcoma. Cancer Res. 2006;66:4742–9.
Dai L, Trillo-Tinoco J, Cao Y, Bonstaff K, Doyle L, Del Valle L, et al. Targeting HGF/c-MET induces cell cycle arrest, DNA damage, and apoptosis for primary effusion lymphoma. Blood. 2015;126:2821–31.
Guan X. Cancer metastases: challenges and opportunities. Acta Pharm Sin B. 2015;5:402–18.
Zambelli A, Biamonti G, Amato A. HGF/c-Met signalling in the tumor microenvironment. Adv Exp Med Biol. 2021;1270:31–44.
Zhang Y, Gao X, Zhu Y, Kadel D, Sun H, Chen J, et al. The dual blockade of MET and VEGFR2 signaling demonstrates pronounced inhibition on tumor growth and metastasis of hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:93.
Ciccarese C, Brunelli M, Montironi R, Fiorentino M, Iacovelli R, Heng D, et al. The prospect of precision therapy for renal cell carcinoma. Cancer Treat Rev. 2016;49:37–44.
Oyama T, Ichimura E, Sano T, Kashiwabara K, Fukuda T, Nakajima T. C-Met expression of thyroid tissue with special reference to papillary carcinoma. Pathol Int. 1998;48:763–8.
Scarpino S, Cancellario d’Alena F, Di Napoli A, Pasquini A, Marzullo A, Ruco LP. Increased expression of Met protein is associated with up-regulation of hypoxia inducible factor-1 (HIF-1) in tumour cells in papillary carcinoma of the thyroid. J Pathol. 2004;202:352–8.
Nakamura Y, Matsubara D, Goto A, Ota S, Sachiko O, Ishikawa S, et al. Constitutive activation of c-Met is correlated with c-Met overexpression and dependent on cell–matrix adhesion in lung adenocarcinoma cell lines. Cancer Sci. 2008;99:14–22.
Charytonowicz E, Cordon-Cardo C, Matushansky I, Ziman M. Alveolar rhabdomyosarcoma: is the cell of origin a mesenchymal stem cell? Cancer Lett. 2009;279:126–36.
Skrzypek K, Kusienicka A, Szewczyk B, Adamus T, Lukasiewicz E, Miekus K, et al. Constitutive activation of MET signaling impairs myogenic differentiation of rhabdomyosarcoma and promotes its development and progression. Oncotarget. 2015;6:31378–98.
Previdi S, Maroni P, Matteucci E, Broggini M, Bendinelli P, Desiderio MA. Interaction between human-breast cancer metastasis and bone microenvironment through activated hepatocyte growth factor/Met and beta-catenin/Wnt pathways. Eur J Cancer. 2010;46:1679–91.
Di Renzo MF, Olivero M, Giacomini A, Porte H, Chastre E, Mirossay L, et al. Overexpression and amplification of the met/HGF receptor gene during the progression of colorectal cancer. Clin Cancer Res. 1995;1:147–54.
Yao JF, Li XJ, Yan LK, He S, Zheng JB, Wang XR, et al. Role of HGF/c-Met in the treatment of colorectal cancer with liver metastasis. J Biochem Mol Toxicol. 2019;33:e22316.
Du F, Li X, Feng W, Qiao C, Chen J, Jiang M, et al. SOX13 promotes colorectal cancer metastasis by transactivating SNAI2 and c-MET. Oncogene. 2020;39:3522–40.
Shen M, Zhao X, Zhao L, Shi L, An S, Huang G, et al. Met is involved in TIGAR-regulated metastasis of non-small-cell lung cancer. Mol Cancer. 2018;17:88.
Michikoshi H, Nakamura T, Sakai K, Suzuki Y, Adachi E, Matsugo S, et al. α-Lipoic acid-induced inhibition of proliferation and met phosphorylation in human non-small cell lung cancer cells. Cancer Lett. 2013;335:472–8.
Miura Y, Kozuki Y, Yagasaki K. Potentiation of invasive activity of hepatoma cells by reactive oxygen species is mediated by autocrine/paracrine loop of hepatocyte growth factor. Biochem Biophys Res Commun. 2003;305:160–5.
Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351–9.
Huang X, Gan G, Wang X, Xu T, Xie W. The HGF-MET axis coordinates liver cancer metabolism and autophagy for chemotherapeutic resistance. Autophagy. 2019;15:1258–79.
Che L, Chi W, Qiao Y, Zhang J, Song X, Liu Y, et al. Cholesterol biosynthesis supports the growth of hepatocarcinoma lesions depleted of fatty acid synthase in mice and humans. Gut. 2020;69:177–86.
Yan B, Jiang Z, Cheng L, Chen K, Zhou C, Sun L, et al. Paracrine HGF/c-MET enhances the stem cell-like potential and glycolysis of pancreatic cancer cells via activation of YAP/HIF-1α. Exp Cell Res. 2018;371:63–71.
Zhang Y, Nguyen TTT, Shang E, Mela A, Humala N, Mahajan A, et al. MET inhibition elicits PGC1α-dependent metabolic reprogramming in glioblastoma. Cancer Res. 2020;80:30–43.
Nassar D, Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu Rev Pathol. 2016;11:47–76.
Saygin C, Matei D, Majeti R, Reizes O, Lathia JD. Targeting cancer stemness in the clinic: from hype to hope. Cell Stem Cell. 2019;24:25–40.
Jeon HM, Lee J. MET: roles in epithelial-mesenchymal transition and cancer stemness. Ann Transl Med. 2017;5:5.
O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10.
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–8.
Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–51.
Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci USA. 2011;108:9951–6.
Vermeulen L, De Sousa EMF, van der Heijden M, Cameron K, de Jong JH, Borovski T, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–76.
Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11.
Xing F, Liu Y, Wu SY, Wu K, Sharma S, Mo YY, et al. Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. 2018;78:4316–30.
Joo KM, Jin J, Kim E, Ho Kim K, Kim Y, Gu Kang B, et al. MET signaling regulates glioblastoma stem cells. Cancer Res. 2012;72:3828–38.
Rath P, Lal B, Ajala O, Li Y, Xia S, Kim J, et al. In vivo c-Met pathway inhibition depletes human glioma xenografts of tumor-propagating stem-like cells. Transl Oncol. 2013;6:104–11.
Flores-Téllez TN, Villa-Treviño S, Piña-Vázquez C. Road to stemness in hepatocellular carcinoma. World J Gastroenterol. 2017;23:6750–76.
Dang H, Steinway SN, Ding W, Rountree CB. Induction of tumor initiation is dependent on CD44s in c-Met+ hepatocellular carcinoma. BMC Cancer. 2015;15:161.
Huang WC, Tung SL, Chen YL, Chen PM, Chu PY. IFI44L is a novel tumor suppressor in human hepatocellular carcinoma affecting cancer stemness, metastasis, and drug resistance via regulating met/Src signaling pathway. BMC Cancer. 2018;18:609.
Peng H, Xue R, Ju Z, Qiu J, Wang J, Yan W, et al. Cancer-associated fibroblasts enhance the chemoresistance of CD73+ hepatocellular carcinoma cancer cells via HGF-Met-ERK1/2 pathway. Ann Transl Med. 2020;8:856.
Jahangiri A, Nguyen A, Chandra A, Sidorov MK, Yagnik G, Rick J, et al. Cross-activating c-Met/β1 integrin complex drives metastasis and invasive resistance in cancer. Proc Natl Acad Sci USA. 2017;114:E8685–94.
Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4.
Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, et al. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell. 2015;162:1271–85.
Zhou L, XD, Sun M, Zhang X, German P, Bai S, et al. Targeting MET and AXL overcomes resistance to sunitinib therapy in renal cell carcinoma. Oncogene. 2016;35:2687–97.
Shojaei F, Lee JH, Simmons BH, Wong A, Esparza CO, Plumlee PA, et al. HGF/c-Met acts as an alternative angiogenic pathway in sunitinib-resistant tumors. Cancer Res. 2010;70:10090–100.
Westover D, Zugazagoitia J, Cho BC, Lovly CM, Paz-Ares L. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol. 2018;29:i10–i19.
Wu YL, Cheng Y, Zhou J, Lu S, Zhang Y, Zhao J, et al. Tepotinib plus gefitinib in patients with EGFR-mutant non-small-cell lung cancer with MET overexpression or MET amplification and acquired resistance to previous EGFR inhibitor (INSIGHT study): an open-label, phase 1b/2, multicentre, randomised trial. Lancet Respir Med. 2020;8:1132–43.
Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–7.
Wang Y, Chen Z, Han X, Li J, Guo H, Shi J. Acquired MET D1228N mutations mediate crizotinib resistance in lung adenocarcinoma with ROS1 fusion: a case report. Oncologist. 2021;26:178–81.
De Bacco F, Luraghi P, Medico E, Reato G, Girolami F, Perera T, et al. Induction of MET by ionizing radiation and its role in radioresistance and invasive growth of cancer. J Natl Cancer Inst. 2011;103:645–61.
Aebersold DM, Landt O, Berthou S, Gruber G, Beer KT, Greiner RH, et al. Prevalence and clinical impact of Met Y1253D-activating point mutation in radiotherapy-treated squamous cell cancer of the oropharynx. Oncogene. 2003;22:8519–23.
Jankowski K, Kucia M, Wysoczynski M, Reca R, Zhao D, Trzyna E, et al. Both hepatocyte growth factor (HGF) and stromal-derived factor-1 regulate the metastatic behavior of human rhabdomyosarcoma cells, but only HGF enhances their resistance to radiochemotherapy. Cancer Res. 2003;63:7926–35.
Kim JH, Kim HS, Kim BJ. Prognostic value of MET copy number gain in non-small-cell lung cancer: an updated meta-analysis. J Cancer. 2018;9:1836–45.
Vuong HG, Ho ATN, Altibi AMA, Nakazawa T, Katoh R, Kondo T. Clinicopathological implications of MET exon 14 mutations in non-small cell lung cancer - a systematic review and meta-analysis. Lung Cancer. 2018;123:76–82.
Pyo JS, Kang G, Cho WJ, Choi SB. Clinicopathological significance and concordance analysis of c-MET immunohistochemistry in non-small cell lung cancers: a meta-analysis. Pathol Res Pr. 2016;212:710–6.
Wang F, Li S, Zhao Y, Yang K, Chen M, Niu H, et al. Predictive role of the overexpression for CXCR4, C-Met, and VEGF-C among breast cancer patients: a meta-analysis. Breast. 2016;28:45–53.
Yan S, Jiao X, Zou H, Li K. Prognostic significance of c-Met in breast cancer: a meta-analysis of 6010 cases. Diagn Pathol. 2015;10:62.
Zagouri F, Bago-Horvath Z, Rössler F, Brandstetter A, Bartsch R, Papadimitriou CA, et al. High MET expression is an adverse prognostic factor in patients with triple-negative breast cancer. Br J Cancer. 2013;108:1100–5.
Liu Y, Yu XF, Zou J, Luo ZH. Prognostic value of c-Met in colorectal cancer: a meta-analysis. World J Gastroenterol. 2015;21:3706–10.
Lee SJ, Lee J, Park SH, Park JO, Lim HY, Kang WK, et al. c-MET overexpression in colorectal cancer: a poor prognostic factor for survival. Clin Colorectal Cancer. 2018;17:165–9.
Choi J, Lee HE, Lee HS, Han N, Kim MA, Kim WH. Evaluation of intratumoral and intertumoral heterogeneity of MET protein expression in gastric cancer. Appl Immunohistochem Mol Morphol. 2018;26:445–45.
Huang KH, Sung IC, Fang WL, Chi CW, Yeh TS, Lee HC, et al. Correlation between HGF/c-Met and Notch1 signaling pathways in human gastric cancer cells. Oncol Rep. 2018;40:294–302.
Catenacci DV, Ang A, Liao WL, Shen J, O’Day E, Loberg RD, et al. MET tyrosine kinase receptor expression and amplification as prognostic biomarkers of survival in gastroesophageal adenocarcinoma. Cancer. 2017;123:1061–70.
Ren JL, Wu HF, Wang WJ, Hu GM, Gu B, Zhang M, et al. C-Met as a potential novel prognostic marker in squamous cell carcinoma and adenocarcinoma of esophagus: evidence from a meta-analysis. Panminerva Med. 2017;59:97–106.
Szturz P, Budíková M, Vermorken JB, Horová I, Gál B, Raymond E, et al. Prognostic value of c-MET in head and neck cancer: a systematic review and meta-analysis of aggregate data. Oral Oncol. 2017;74:68–76.
Vsiansky V, Gumulec J, Raudenska M, Masarik M. Prognostic role of c-Met in head and neck squamous cell cancer tissues: a meta-analysis. Sci Rep. 2018;8:10370.
Kim JH, Kim HS, Kim BJ, Lee J, Jang HJ. Prognostic value of c-Met overexpression in pancreatic adenocarcinoma: a meta-analysis. Oncotarget. 2017;8:73098–104.
Kim JH, Kim HS, Kim BJ, Jang HJ, Lee J. Prognostic value of c-Met overexpression in hepatocellular carcinoma: a meta-analysis and review. Oncotarget. 2017;8:90351–7.
Kim JH, Kim BJ, Kim HS. Clinicopathological impacts of high c-Met expression in renal cell carcinoma: a meta-analysis and review. Oncotarget. 2017;8:75478–87.
Gibney GT, Aziz SA, Camp RL, Conrad P, Schwartz BE, Chen CR, et al. c-Met is a prognostic marker and potential therapeutic target in clear cell renal cell carcinoma. Ann Oncol. 2013;24:343–9.
Schiefer AI, Mesteri I, Berghoff AS, Haitel A, Schmidinger M, Preusser M, et al. Evaluation of tyrosine kinase receptors in brain metastases of clear cell renal cell carcinoma reveals c-Met as a negative prognostic factor. Histopathology. 2015;67:799–805.
Peng J, Qi S, Wang P, Li W, Liu C, Li F. Diagnosis and prognostic significance of c-Met in cervical cancer: a meta-analysis. Dis Markers. 2016;2016:6594016.
Lux A, Kahlert C, Grützmann R, Pilarsky C. c-Met and PD-L1 on circulating exosomes as diagnostic and prognostic markers for pancreatic cancer. Int J Mol Sci. 2019;20:3305.
Tsuji T, Sakamori Y, Ozasa H, Yagi Y, Ajimizu H, Yasuda Y, et al. Clinical impact of high serum hepatocyte growth factor in advanced non-small cell lung cancer. Oncotarget. 2017;8:71805–16.
Park DJ, Yoon C, Thomas N, Ku GY, Janjigian YY, Kelsen DP, et al. Prognostic significance of targetable angiogenic and growth factors in patients undergoing resection for gastric and gastroesophageal junction cancers. Ann Surgical Oncol. 2014;21:1130–7.
Zhang Y, Wu JZ, Yang YQ, Ma R, Zhang JY, Feng JF. Expression of growth‑regulated oncogene‑1, hepatocyte growth factor, platelet‑derived growth factor‑AA and soluble E‑selectin and their association with high‑risk human papillomavirus infection in squamous cell carcinoma of the uterine cervix. Mol Med Rep. 2014;10:1013–24.
Liang QL, Mo ZY, Wang P, Li X, Liu ZX, Zhou ZM. The clinical value of serum hepatocyte growth factor levels in patients undergoing primary radiotherapy for glioma: effect on progression-free survival. Med Oncol. 2014;31:122.
Kubo Y, Fukushima S, Inamori Y, Tsuruta M, Egashira S, Yamada-Kanazawa S, et al. Serum concentrations of HGF are correlated with response to anti-PD-1 antibody therapy in patients with metastatic melanoma. J Dermatol Sci. 2019;93:33–40.
Saltarella I, Morabito F, Giuliani N, Terragna C, Omedè P, Palumbo A, et al. Prognostic or predictive value of circulating cytokines and angiogenic factors for initial treatment of multiple myeloma in the GIMEMA MM0305 randomized controlled trial. J Hematol Oncol. 2019;12:4.
Gupta A, Karakiewicz PI, Roehrborn CG, Lotan Y, Zlotta AR, Shariat SF. Predictive value of plasma hepatocyte growth factor/scatter factor levels in patients with clinically localized prostate cancer. Clin Cancer Res. 2008;14:7385–90.
Kim H, Youk J, Yang Y, Kim TY, Min A, Ham HS, et al. Prognostic implication of serum hepatocyte growth factor in stage II/III breast cancer patients who received neoadjuvant chemotherapy. J Cancer Res Clin Oncol. 2016;142:707–14.
van den Bent M, Azaro A, De Vos F, Sepulveda J, Yung WKA, Wen PY, et al. A phase Ib/II, open-label, multicenter study of INC280 (capmatinib) alone and in combination with buparlisib (BKM120) in adult patients with recurrent glioblastoma. J Neurooncol. 2020;146:79–89.
Wu YL, Zhang L, Kim DW, Liu X, Lee DH, Yang JC, et al. Phase Ib/II study of capmatinib (INC280) plus gefitinib after failure of epidermal growth factor receptor (EGFR) inhibitor therapy in patients with EGFR-mutated, MET factor-dysregulated non–small-cell lung cancer. J Clin Oncol. 2018;36:3101–9.
Wolf J, Seto T, Han JY, Reguart N, Garon EB, Groen HJM, et al. Capmatinib in MET exon 14-mutated or MET-amplified non-small-cell lung cancer. Engl J Med. 2020;383:944–57.
Moro-Sibilot D, Cozic N, Pérol M, Mazières J, Otto J, Souquet PJ, et al. Crizotinib in c-MET- or ROS1-positive NSCLC: results of the AcSé phase II trial. Ann Oncol. 2019;30:1985–91.
Aparicio T, Cozic N, de la Fouchardière C, Meriaux E, Plaza J, Mineur L. et al.The activity of crizotinib in chemo-refractory MET-amplified esophageal and gastric adenocarcinomas: results from the AcSé-crizotinib program.Target Oncol . 2021;16:381–8.
Ou SH, Kwak EL, Siwak-Tapp C, Dy J, Bergethon K, Clark JW, et al. Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol. 2011;6:942–6.
Drilon A, Clark JW, Weiss J, Ou SI, Camidge DR, Solomon, et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat Med. 2020;26:47–51.
Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó L, Ahn MJ, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N. Engl J Med. 2013;368:2385–94.
Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl J Med. 2014;371:2167–77.
Solomon BJ, Kim DW, Wu YL, Nakagawa K, Mekhail T, Felip E, et al. Final overall survival analysis from a study comparing first-line crizotinib versus chemotherapy in ALK-mutation-positive non-small-cell lung cancer. J Clin Oncol. 2018;36:2251–8.
Wu YL, Lu S, Lu Y, Zhou J, Shi YK, Sriuranpong V, et al. Results of Profile 1029, a phase III comparison of first- line crizotinib versus chemotherapy in East Asian patients with ALK-positive advanced non-small cell lung cancer. J Thorac Oncol. 2018;13:1539–48.
Paik PK, Felip E, Veillon R, Sakai H, Cortot AB, Garassino MC, et al. Tepotinib in non-small-cell lung cancer with MET exon 14 skipping mutations. N. Engl J Med. 2020;383:931–43.
Burbridge MF, Bossard CJ, Saunier C, Fejes I, Bruno A, Leonce S, et al. S49076 is a novel kinase inhibitor of MET, AXL, and FGFR with strong preclinical activity alone and in association with bevacizumab. Mol Cancer Ther. 2013;12:1749–62.
Rodon J, Postel-Vinay S, Hollebecque A, Nuciforo P, Azaro A, Cattan V, et al. First-in-human phase I study of oral S49076, a unique MET/AXL/FGFR inhibitor, in advanced solid tumours. Eur J Cancer. 2017;81:142–50.
Bertran-Alamillo J, Cattan V, Schoumacher M, Codony-Servat J, Giménez-Capitán A, Cantero F, et al. AURKB as a target in non-small cell lung cancer with acquired resistance to anti-EGFR therapy. Nat Commun. 2019;10:1812.
Park K, Chang GC, Curigliano G, Lim WT, Soo RA, Molina-Vila MA, et al. Phase I results of S49076 plus gefitinib in patients with EGFR TKI-resistant non-small cell lung cancer harbouring MET/AXL dysregulation. Lung Cancer. 2021;155:127–35.
Kurzrock R, Sherman SI, Ball DW, Forastiere AA, Cohen RB, Mehra R, et al. Activity of XL184 (Cabozantinib), an oral tyrosine kinase inhibitor, in patients with medullary thyroid cancer. J Clin Oncol. 2011;29:2660–6.
Smith DC, Smith MR, Sweeney C, Elfiky AA, Logothetis C, Corn PG, et al. Cabozantinib in patients with advanced prostate cancer: results of a phase II randomized discontinuation trial. J Clin Oncol. 2013;31:412–9.
Kelley RK, Verslype C, Cohn AL, Yang TS, Su WC, Burris H, et al. Cabozantinib in hepatocellular carcinoma: results of a phase 2 placebo-controlled randomized discontinuation study. Ann Oncol. 2017;28:528–34.
Wakelee HA, Gettinger S, Engelman J, Jänne PA, West H, Subramaniam DS, et al. A phase Ib/II study of cabozantinib (XL184) with or without erlotinib in patients with non-small cell lung cancer. Cancer Chemother Pharm. 2017;79:923–32.
Choueiri TK, Halabi S, Sanford BL, Hahn O, Michaelson MD, Walsh MK, et al. Cabozantinib versus sunitinib as initial targeted therapy for patients with metastatic renal cell carcinoma of poor or intermediate risk: the alliance A031203 CABOSUN trial. J Clin Oncol. 2017;35:591–7.
Choueiri TK, Escudier B, Powles T, Mainwaring PN, Rini BI, Donskov F, et al. Cabozantinib versus everolimus in advanced renal-cell carcinoma. N. Engl J Med. 2015;373:1814–23.
Choueiri TK, Escudier B, Powles T, Tannir NM, Mainwaring PN, Rini BI, et al. Cabozantinib versus everolimus in advanced renal cell carcinoma (METEOR): final results from a randomised, open-label, phase 3 trial. Lancet Oncol. 2016;17:917–27.
Hofmann F, Hwang EC, Lam TB, Bex A, Yuan Y, Marconi LS, et al. Targeted therapy for metastatic renal cell carcinoma. Cochrane Database Syst Rev. 2020;10:CD012796.
Choueiri TK, Albiges L, Powles T, Scheffold C, Wang F, Motzer R. A phase III study (COSMIC-313) of cabozantinib (C) in combination with nivolumab (N) and ipilimumab (I) in patients (pts) with previously untreated advanced renal cell carcinoma (aRCC) of intermediate or poor risk. J Clin Oncol. 2020;38:TPS767.
Smith M, De Bono J, Sternberg C, Le Moulec S, Oudard S, De Giorgi U, et al. Phase III study of cabozantinib in previously treated metastatic castration-resistant prostate cancer: COMET-1. J Clin Oncol. 2016;34:3005–13.
Abou-Alfa GK, Meyer T, Cheng AL, El-Khoueiry AB, Rimassa L, Ryoo BY, et al. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N Engl J Med. 2018;379:54–63.
Schlumberger M, Elisei R, Müller S, Schöffski P, Brose M, Shah M, et al. Overall survival analysis of EXAM, a phase III trial of cabozantinib in patients with radiographically progressive medullary thyroid carcinoma. Ann Oncol. 2017;28:2813–9.
Engstrom LD, Aranda R, Lee M, Tovar EA, Essenburg CJ, Madaj Z, et al. Glesatinib exhibits antitumor activity in lung cancer models and patients harboring MET exon 14 mutations and overcomes mutation-mediated resistance to type I MET inhibitors in nonclinical models. Clin Cancer Res. 2017;23:6661–72.
Roohullah A, Cooper A, Lomax AJ, Aung J, Barge A, Chow L, et al. A phase I trial to determine safety and pharmacokinetics of ASLAN002, an oral MET superfamily kinase inhibitor, in patients with advanced or metastatic solid cancers. Invest New Drugs. 2018;36:886–94.
Doi T, Matsubara N, Kawai A, Naka N, Takahashi S, Uemura H, et al. Phase I study of TAS-115, a novel oral multi-kinase inhibitor, in patients with advanced solid tumors. Invest New Drugs. 2020;38:1175–85.
Penuel E, Li C, Parab V, Burton L, Cowan KJ, Merchant M, et al. HGF as a circulating biomarker of onartuzumab treatment in patients with advanced solid tumors. Mol Cancer Ther. 2013;12:1122–30.
Cui JJ, Tran-Dubé M, Shen H, Nambu M, Kung PP, Pairish M, et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J Med Chem. 2011;54:6342–63.
Drilon AE, Camidge DR, Ou S-HI, Clark JW, Socinski MA, Weiss J, et al. Efficacy and safety of crizotinib in patients (pts) with advanced MET exon 14-altered non-small cell lung cancer (NSCLC). J Clin Oncol. 2016;34:108.
Puccini A, Marín-Ramos NI, Bergamo F, Schirripa M, Lonardi S, Lenz HJ, et al. Safety and tolerability of c-MET Inhibitors in cancer. Drug Saf. 2019;42:211–33.
Baltschukat S, Engstler BS, Huang A, Hao HX, Tam A, Wang HQ, et al. Capmatinib (INC280) is active against models of non–small cell lung cancer and other cancer types with defined mechanisms of MET activation. Clin Cancer Res. 2019;25:3164–75.
Marona P, Górka J, Kotlinowski J, Majka M, Jura J, Miekus K. C-Met as a key factor responsible for sustaining undifferentiated phenotype and therapy resistance in renal carcinomas. Cells. 2019;8:272.
Li A, Yang JJ, Zhang XC, Zhang Z, Su J, Gou LY, et al. Acquired MET Y1248H and D1246N mutations mediate resistance to MET inhibitors in non-small cell lung cancer. Clin Cancer Res. 2017;23:4929–37.
Recondo G, Bahcall M, Spurr LF, Che J, Ricciuti B, Leonardi GC, et al. Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET exon 14-mutant NSCLC. Clin Cancer Res. 2020;26:2615–25.
Li H, Li CW, Li X, Ding Q, Guo L, Liu S, et al. MET inhibitors promote liver tumor evasion of the immune response by stabilizing PDL1. Gastroenterology. 2019;156:1849–.e1813.
Maulik G, Kijima T, Ma PC, Ghosh SK, Lin J, Shapiro GI, et al. Modulation of the c-Met/hepatocyte growth factor pathway in small cell lung cancer. Clin Cancer Res. 2002;8:620–7.
Antonia SJ, López-Martin JA, Bendell J, Ott PA, Taylor M, Eder JP, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol. 2016;17:883–95.
Ready N, Farago AF, de Braud F, Atmaca A, Hellmann MD, Schneider JG, et al. Third- line nivolumab monotherapy in recurrent SCLC: CheckMate 032. J Thorac Oncol. 2019;14:237–44.
Acknowledgements
J.P. designed the review, wrote, and revised the manuscript with input from J.F., X.S., Z.L., L.D., X.L., X.F., and J.P. The literature search was performed and analyzed by J.F., X.S., Z.L., L.D., X.L., X.F., and J.P. The figures of the review were mainly painted by J.F., X.F., L.D., and X.L. A major part of Table 1 was completed by J.P. and J.F.; X.F. participated in revising the review critically for the important intellectual content. All authors have read and approved the manuscript.
Funding
This work has been supported by the National Natural Science Foundation of China (No. 81602631), the Guangdong Natural Science Foundation of China (No. 2016A030310280), the Students’ Laboratory Open Project of Guangzhou Medical University (No. C195015021), 2017 Elite Talents Training Plan of the Third Affiliated Hospital of Guangzhou Medical University (J.P.), and Special Funds for the Cultivation of Guangdong College Students’ Scientific and Technological Innovation: “Climbing Program” Special Funds (No. pdjh2020b0490).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fu, J., Su, X., Li, Z. et al. HGF/c-MET pathway in cancer: from molecular characterization to clinical evidence. Oncogene 40, 4625–4651 (2021). https://doi.org/10.1038/s41388-021-01863-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01863-w