Abstract
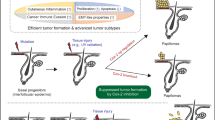
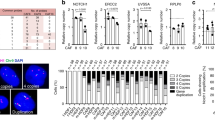
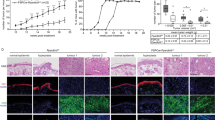
Cutaneous squamous cell carcinoma (cSCC) ranks second in the frequency of all skin cancers. The balance between keratinocyte proliferation and differentiation is disrupted in the pathological development of cSCC. DLX3 is a homeobox transcription factor which plays pivotal roles in embryonic development and epidermal homeostasis. To investigate the impact of DLX3 expression on cSCC prognosis, we carried out clinicopathologic analysis of DLX3 expression which showed statistical correlation between tumors of higher pathologic grade and levels of DLX3 protein expression. Further, Kaplan–Meier survival curve analysis demonstrated that low DLX3 expression correlated with poor patient survival. To model the function of Dlx3 in skin tumorigenesis, a two-stage dimethylbenzanthracene (DMBA)/12-O-tetradecanoylphorbol 13-acetate (TPA) study was performed on mice genetically depleted of Dlx3 in skin epithelium (Dlx3cKO). Dlx3cKO mice developed significantly more tumors, with more rapid tumorigenesis compared to control mice. In Dlx3cKO mice treated only with DMBA, tumors developed after ~16 weeks suggesting that loss of Dlx3 has a tumor promoting effect. Whole transcriptome analysis of tumor and skin tissue from our mouse model revealed spontaneous activation of the EGFR–ERBB2 pathway in the absence of Dlx3. Together, our findings from human and mouse model system support a tumor suppressive function for DLX3 in skin and underscore the efficacy of therapeutic approaches that target EGFR–ERBB2 pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Que SK, Zwald F, Schmults C. Cutaneous squamous cell carcinoma: incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol. 2018;78:237–47.
Karia PS, Han J, Schmults CD. Cutaneous squamous cell carcinoma: estimated incidence of disease, nodal metastastasis, and deaths from disease in the United States, 2012. J Am Acad Dermatol. 2013;68:957–66.
Lallas A, Pyne J, Kyrgidis A, Andreani S, Argenziano G, Cavaller A, et al. The clinical and dermoscopic features of invasive cutaneous squamous cell carcinoma depend on the histopathological grade of differentiation. Br J Dermatol. 2015;172:1308–15.
Cillo C, Faiella A, Cantile M, Boncinelli E. Homeobox genes and cancer. Exp Cell Res. 1999;248:1–9.
Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–85.
Sun Y, Zhou B, Mao F, Xu J, Miao H, Zou Z, et al. HOXA9 reprograms the enhancer landscape to promote leukemogenesis. Cancer Cell. 2018;34:643–58.
Shah N, Sukumar S. The hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–71.
Samuel S, Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur J Cancer. 2005;41:2428–37.
Luo Z, Rhie SK, Farnham PJ. The enigmatic hox genes: can we crack their code? Cancers. 2019;11:323.
Hwang J, Mehrani T, Millar SE, Morasso MI. Dlx3 is a crucial regulator of hair follicle differentiation and cycling. Development. 2008;135:3149–59.
Morasso MI, Radoja N. Dlx genes, p63, and ectodermal dysplasias. Birth Defects Res C Embryo Today. 2005;75:163–71.
Hwang J, Kita R, Kwon HS, Choi EH, Lee SH, Udey MC, et al. Epidermal ablation of Dlx3 is linked to IL-17-associated skin inflammation. PNAS. 2011;108:11566–71.
Bhattacharya S, Kim JC, Ogawa Y, Nakato G, Nagle V, Brooks SR, et al. DLX3-dependent STAT3 signaling in keratinocytes regulates skin immune homeostasis. J Investig Dermatol. 2018;138:1052–61.
Palazzo E, Kellett M, Cataisson C, Gormley A, Bible PW, Pietroni V, et al. The homeoprotein DLX3 and tumor suppressor p53 co-regulate cell cycle progression and squamous tumor growth. Oncogene. 2016;35:3114.
Witsch E, Sela M, Yarden Y. Roles for growth factors in cancer progression. Physiology. 2010;25:85–101.
Dahlhoff M, Muzumdar S, Schafer M, Schneider MR. ERBB2 is essential for the growth of chemically induced skin tumors in mice. J Investig Dermatol. 2017;137:921–30.
Dahlhoff M, Schafer M, Muzumdar S, Rose C, Schneider MR. ERBB3 is required for tumor promotion in a mouse model of skin carcinogenesis. Mol Oncol. 2015;9:1825–33.
Chan KS, Carbajal S, Kiguchi K, Clifford J, Sano S, DiGiovanni J. Epidermal growth factor receptor‐mediated activation of Stat3 during multistage skin carcinogenesis. Cancer Res. 2004;64:2382–9.
Dahlhoff M, Rose C, Wolf E, Schneider MR. Decreased incidence of papillomas in mice with impaired EGFR function during multi‐stage skin carcinogenesis. Exp Dermatol. 2011;20:290–3.
Subramanian J, Katta A, Masood A, Vudem DR, Kancha RK. Emergence of ERBB2 mutation as a biomarker and an actionable target in solid cancers. Oncologist. 2019;24:e1303–14.
Kiguchi K, Kitamura T, Moore T, Rumi M, Chang HC, Treece D, et al. Dual inhibition of the epidermal growth factor receptor and erbB2 effectively inhibits the promotion of skin tumors during two-stage carcinogenesis. Cancer Prev Res. 2010;3:940–52.
Abel EL, Angel JM, Kiguchi K, DiGiovanni J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat Protoc. 2009;4:1350–62.
Darwiche N, Ryscavage A, Perez-Lorenzo R, Wright L, Bae D-S, Hennings H, et al. Expression profile of skin papillomas with high cancer risk displays a unique genetic signature that clusters with squamous cell carcinomas and predicts risk for malignant conversion. Oncogene. 2007;26:6885–95.
Palazzo E, Kellett MD, Cataisson C, Bible PW, Bhattacharya S, Sun HW, et al. A novel DLX3-PKC integrated signaling network drives keratinocyte differentiation. Cell Death Differ. 2017;4:717–30.
Plowright L, Harrington K, Pandha H, Morgan R. HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer (targeting HOX genes in lung cancer). Br J Cancer. 2009;100:470–5.
Gilbert PM, Mouw JK, Unger MA, Lakins JN, Gbegnon MK, Clemmer VB, et al. Hoxa9 regulates brca1 expression to modulate human breast tumor phenotype. J Clin Investig. 2010;120:1535–50.
Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, et al. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–8.
Wheeler DL, Verma AK, Denning MF. Mouse models of the skin: models to define mechanisms of skin carcinogenesis. J Skin Cancer. 2013:971495.
Wilker E, Lu J, Rho O, Carbajal S, Beltrán L, DiGiovanni J. Role of PI3K/Akt signaling in insulin-like growth factor-1 (IGF-1) skin tumor promotion. Mol Carcinog. 2005;44:137–45.
Cataisson C, Ohman R, Patel G, Pearson A, Tsien M, Jay S, et al. Inducible cutaneous inflammation reveals a protumorigenic role for keratinocyte CXCR2 in skin carcinogenesis. Cancer Res. 2009;69:319–28.
Mueller MM. Inflammation in epithelial skin tumours: old stories and new ideas. Eur J Cancer. 2006;42:735–44.
Gimenez-Conti I, Aldaz CM, Bianchi AB, Roop DR, Slaga TJ, Conti CJ. Early expression of type I K13 keratin in the progression of mouse skin papillomas. Carcinogenesis. 1990;11:1995–9.
Toftgard R, Yuspa SH, Roop DR. Keratin gene expression in mouse skin tumors and in mouse skin treated with 12-Otetradecanoylphorbol-13-acetate. Cancer Res. 1985;45:5845–50.
Nischt R, Roop DR, Mehrel T, Yuspa SH, Rentrop M, Winter H, et al. Aberrant expression during two-stage mouse skin carcinogenesis of a type I 47-kDa keratin, K13, normally associated with terminal differentiation of internal stratified epithelia. Mol Carcinog. 1988;1:96–108.
Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–7.
Nyati MK, Morgan MA, Feng FY, Lawrence TS. Integration of EGFR inhibitors with radiochemotherapy [published correction appears in Nat Rev Cancer. 2006 Dec;6(12):974]. Nat Rev Cancer. 2006;6:876–85.
Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315:683–96.
Herbst RS, Shin DM. Monoclonal antibodies to target epidermal growth factor receptor-positive tumors: a new paradigm for cancer therapy. Cancer. 2002;94:1593–611.
Kiguchi K, Beltrán L, Rupp T, DiGiovanni J. Altered expression of epidermal growth factor receptor ligands in tumor promoter-treated mouse epidermis and in primary mouse skin tumors induced by an initiation-promotion protocol. Mol Carcinog. 1998;22:73–83.
El-Abaseri TB, Fuhrman J, Trempus C, Shendrik I, Tennant RW, Hansen LA. Chemoprevention of UV light-induced skin tumorigenesis by inhibition of the epidermal growth factor receptor. Cancer Res. 2005;65:3958–65.
Xian W, Rosenberg MP, DiGiovanni J. Activation of erbb2 and c-src in phorbol ester-treated mouse epidermis: possible role in mouse skin tumor promotion. Oncogene. 1997;14:1435–44.
Schneider M, Yarden Y. The EGFR-HER2 module: a stem cell approach to understanding a prime target and driver of solid tumors. Oncogene. 2016;35:2949–60.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8.
Lichti U, Anders J, Yuspa SH. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protoc. 2008;3:799–810.
Acknowledgements
This work was supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases, NIH (ZIA-AR041124 to MIM). We thank Gutierrez-Cruz and S. Dell’Orso of the NIAMS Genome Analysis Core Facility and members of the NIAMS Light Imaging Core Facility. This work used the computational resources of the NIH High-Performance Computing Biowulf Cluster. We also thank all members of our laboratories for their continuous support. BioRender was used to create experimental design schematic.
Author information
Authors and Affiliations
Contributions
DB, SM, CC, AU, and MIM conceptualize the work; DB, SM, CC, AU, and MIM designed research; DB, SM, CC, AU, YI, MK, EP, and KH performed research; DB, SM, CC, AU, YI, AS, AO, SRB, MK, EP, SM, SY, and MIM analyzed data; and DB, SM, and MIM wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Bajpai, D., Mehdizadeh, S., Uchiyama, A. et al. Loss of DLX3 tumor suppressive function promotes progression of SCC through EGFR–ERBB2 pathway. Oncogene 40, 3680–3694 (2021). https://doi.org/10.1038/s41388-021-01802-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01802-9
This article is cited by
-
CD271 activation prevents low to high-risk progression of cutaneous squamous cell carcinoma and improves therapy outcomes
Journal of Experimental & Clinical Cancer Research (2023)