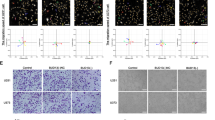
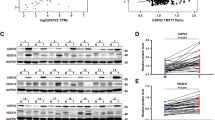
Abstract
Tumor angiogenesis plays vital roles in tumorigenesis and development; regulatory mechanism of angiogenesis is still not been fully elucidated. NSD2, a histone methyltransferase catalyzing di-methylation of histone H3 at lysine 36, has been proved a critical molecule in proliferation, metastasis, and tumorigenesis. But its role in tumor angiogenesis remains unknown. Here we demonstrated that NSD2 promoted tumor angiogenesis in vitro and in vivo. Furthermore, we confirmed that the angiogenic function of NSD2 was mediated by STAT3. Momentously, we found that NSD2 promoted the methylation and activation of STAT3. In addition, mass spectrometry and site-directed mutagenesis assays revealed that NSD2 methylated STAT3 at lysine 163 (K163). Meanwhile, K to R mutant at K163 of STAT3 attenuated the activation and angiogenic function of STAT3. Taken together, we conclude that methylation of STAT3 catalyzed by NSD2 promotes the activation of STAT3 pathway and enhances the ability of tumor angiogenesis. Our findings investigate a NSD2-dependent methylation–phosphorylation regulation pattern of STAT3 and reveal that NSD2/STAT3/VEGFA axis might be a potential target for tumor therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) with the dataset identifier PXD021336.
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol cell Biol. 2007;8:464–78.
De Palma M, Biziato D, Petrova TV. Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer. 2017;17:457–74.
Canavese M, Ngo DT, Maddern GJ, Hardingham JE, Price TJ, Hauben E. Biology and therapeutic implications of VEGF-A splice isoforms and single-nucleotide polymorphisms in colorectal cancer. Int J Cancer. 2017;140:2183–91.
Stratman AN, Schwindt AE, Malotte KM, Davis GE. Endothelial-derived PDGF-BB and HB-EGF coordinately regulate pericyte recruitment during vasculogenic tube assembly and stabilization. Blood. 2010;116:4720–30.
Stratman AN, Davis MJ, Davis GE. VEGF and FGF prime vascular tube morphogenesis and sprouting directed by hematopoietic stem cell cytokines. Blood. 2011;117:3709–19.
Saltz LB. Bevacizumab in colorectal cancer: it should have worked. Lancet Oncol. 2016;17:1469–70.
Smeets D, Miller IS, O’Connor DP, Das S, Moran B, Boeckx B, et al. Copy number load predicts outcome of metastatic colorectal cancer patients receiving bevacizumab combination therapy. Nat Commun. 2018;9:4112.
Haibe Y, Kreidieh M, El Hajj H, Khalifeh I, Mukherji D, Temraz S, et al. Resistance mechanisms to anti-angiogenic therapies in cancer. Front Oncol. 2020;10:221.
Darnell JE Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–21.
Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, et al. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc Natl Acad Sci USA. 1996;93:3963–6.
Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–46.
Yu H, Jove R. The STATs of cancer-new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105.
Bharadwaj U, Kasembeli MM, Robinson P, Tweardy DJ. Targeting Janus kinases and signal transducer and activator of transcription 3 to treat inflammation, fibrosis, and cancer: rationale, progress, and caution. Pharmacol Rev. 2020;72:486–526.
Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA. 2010;107:21499–504.
Dasgupta M, Dermawan JK, Willard B, Stark GR. STAT3-driven transcription depends upon the dimethylation of K49 by EZH2. Proc Natl Acad Sci USA. 2015;112:3985–90.
Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–52.
Luo J, Wang K, Yeh S, Sun Y, Liang L, Xiao Y, et al. LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat Commun. 2019;10:2571.
Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–73.
Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44:609–20.
Stec I, Wright TJ, van Ommen GJ, de Boer PA, van Haeringen A, Moorman AF, et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–82.
Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–91.
Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7:585–98.
Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–34.
Keats JJ, Maxwell CA, Taylor BJ, Hendzel MJ, Chesi M, Bergsagel PL, et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood. 2005;105:4060–9.
Foltz SM, Gao Q, Yoon CJ, Sun H, Yao L, Li Y, et al. Evolution and structure of clinically relevant gene fusions in multiple myeloma. Nat Commun. 2020;11:2666.
Zhang J, Lee YR, Dang F, Gan W, Menon AV, Katon JM, et al. PTEN methylation by NSD2 controls cellular sensitivity to DNA damage. Cancer Discov. 2019;9:1306–23.
Hudlebusch HR, Santoni-Rugiu E, Simon R, Ralfkiær E, Rossing HH, Johansen JV, et al. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res. 2011;17:2919–33.
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58.
Stubbs M, Burn T, Sparks R, Maduskuie T, Diamond S, Rupar M. et al. The novel bromodomain and extraterminal domain inhibitor INCB054329 induces vulnerabilities in myeloma cells that inform rational combination strategies. Clin Cancer Res. 2019;25:300–11.
Biggar KK, Li SS. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 2015;16:5–17.
Zhang X, Huang Y, Shi X. Emerging roles of lysine methylation on non-histone proteins. Cell Mol Life Sci. 2015;72:4257–72.
Hamamoto R, Saloura V, Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer. 2015;15:110–24.
Wang G, Long J, Gao Y, Zhang W, Han F, Xu C, et al. SETDB1-mediated methylation of Akt promotes its K63-linked ubiquitination and activation leading to tumorigenesis. Nat Cell Biol. 2019;21:214–25.
Guo J, Dai X, Laurent B, Zheng N, Gan W, Zhang J, et al. AKT methylation by SETDB1 promotes AKT kinase activity and oncogenic functions. Nat cell Biol. 2019;21:226–37.
Hou Z, Sun L, Xu F, Hu F, Lan J, Song D, et al. Blocking histone methyltransferase SETDB1 inhibits tumorigenesis and enhances cetuximab sensitivity in colorectal cancer. Cancer Lett. 2020;487:63–73.
Park JW, Chae YC, Kim JY, Oh H, Seo SB. Methylation of Aurora kinase A by MMSET reduces p53 stability and regulates cell proliferation and apoptosis. Oncogene. 2018;37:6212–24.
Lhoumaud P, Badri S, Rodriguez-Hernaez J, Sakellaropoulos T, Sethia G, Kloetgen A, et al. NSD2 overexpression drives clustered chromatin and transcriptional changes in a subset of insulated domains. Nat Commun. 2019;10:4843.
Cheong CM, Mrozik KM, Hewett DR, Bell E, Panagopoulos V, Noll JE, et al. Twist-1 is upregulated by NSD2 and contributes to tumour dissemination and an epithelial-mesenchymal transition-like gene expression signature in t(4;14)-positive multiple myeloma. Cancer Lett. 2020;475:99–108.
He C, Liu C, Wang L, Sun Y, Jiang Y, Hao Y. Histone methyltransferase NSD2 regulates apoptosis and chemosensitivity in osteosarcoma. Cell Death Dis. 2019;10:65.
Wang JJ, Zou JX, Wang H, Duan ZJ, Wang HB, Chen P, et al. Histone methyltransferase NSD2 mediates the survival and invasion of triple-negative breast cancer cells via stimulating ADAM9-EGFR-AKT signaling. Acta Pharmacologica Sin. 2019;40:1067–75.
Aytes A, Giacobbe A, Mitrofanova A, Ruggero K, Cyrta J, Arriaga J, et al. NSD2 is a conserved driver of metastatic prostate cancer progression. Nat Commun. 2018;9:5201.
Xie Z, Chooi JY, Toh SHM, Yang D, Basri NB, Ho YS, et al. MMSET I acts as an oncoprotein and regulates GLO1 expression in t(4;14) multiple myeloma cells. Leukemia. 2019;33:739–48.
Swaroop A, Oyer JA, Will CM, Huang X, Yu W, Troche C, et al. An activating mutation of the NSD2 histone methyltransferase drives oncogenic reprogramming in acute lymphocytic leukemia. Oncogene. 2019;38:671–86.
Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40:D1144–9. Database issue.
Acknowledgements
We are grateful to the members in Guihua Wang’s lab and Junbo Hu’s lab for the critical inputs and suggestions. This work is supported by NSFC (No. 81773113 GW, No. 81922053 GW, No.81702264 XC, No. 81974432 GW, and No. 81874186 JH).
Author information
Authors and Affiliations
Contributions
GW conceived the project. GW and JH acquired funding and designed the majority of experiments. ZC and XL supervised the project and gave some advice. DS wrote the manuscript and performed most of the molecular biological experiments. JL analyzed the results. YC, AL, and QW performed most of the phenotype experiments. CZ did the mass spectrometry detection and analysis. YF and JW made their efforts in the bioinformatics analysis. XC made contributions and provided support in the process of revision.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval and consent to participate
All patient specimens mentioned in this study were approved by the Ethics Committee of Tongji Hospital following the Declaration of Helsinki and informed consents were signed before the operation. Animal experiments were performed strictly following the Animal Study Guideline of Huazhong University of Science and Technology.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Song, D., Lan, J., Chen, Y. et al. NSD2 promotes tumor angiogenesis through methylating and activating STAT3 protein. Oncogene 40, 2952–2967 (2021). https://doi.org/10.1038/s41388-021-01747-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01747-z
This article is cited by
-
METTL3 and STAT3 form a positive feedback loop to promote cell metastasis in hepatocellular carcinoma
Cell Communication and Signaling (2023)
-
PRMT5 methylating SMAD4 activates TGF-β signaling and promotes colorectal cancer metastasis
Oncogene (2023)
-
Comprehensive pan-cancer analysis of STAT3 as a prognostic and immunological biomarker
Scientific Reports (2023)
-
Loss of Histone Methyltransferase KMT2D Attenuates Angiogenesis in the Ischemic Heart by Inhibiting the Transcriptional Activation of VEGF-A
Journal of Cardiovascular Translational Research (2023)
-
Role of chemokine-mediated angiogenesis in resistance towards crizotinib and its reversal by anlotinib in EML4-ALK positive NSCLC
Journal of Translational Medicine (2022)